1BZ3
 
 | |
6BQK
 
 | |
6BQJ
 
 | |
1UNC
 
 | | Solution structure of the human villin C-terminal headpiece subdomain | | Descriptor: | VILLIN 1 | | Authors: | Vermeulen, W, Van Troys, M, Vanhaesebrouck, P, Verschueren, M, Fant, F, Ampe, C, Martins, J, Borremans, F. | | Deposit date: | 2003-09-09 | | Release date: | 2004-07-15 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the C-Terminal Headpiece Subdomains of Human Villin and Advillin, Evaluation of Headpiece F-Actin-Binding Requirements
Protein Sci., 13, 2004
|
|
4Z0X
 
 | |
5UJ2
 
 | | Crystal structure of HCV NS5B genotype 2A JFH-1 isolate with S15G E86Q E87Q C223H V321I mutations and Delta8 neta hairpoin loop deletion in complex with GS-639476 (diphsohate version of GS-9813), Mn2+ and symmetrical primer template 5'-AUAAAUUU | | Descriptor: | (1S)-1-(4-aminoimidazo[2,1-f][1,2,4]triazin-7-yl)-1,4-anhydro-2-deoxy-2-fluoro-5-O-[(S)-hydroxy(phosphonooxy)phosphoryl]-2-methyl-D-ribitol, CHLORIDE ION, Genome polyprotein, ... | | Authors: | Edwards, T.E, Fox III, D, Appleby, T.C, Murakami, E, Rey, A, McGrath, M.E. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a 2'-fluoro-2'-C-methyl C-nucleotide HCV polymerase inhibitor and a phosphoramidate prodrug with favorable properties.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
2KT6
 
 | | Structural homology between the C-terminal domain of the PapC usher and its plug | | Descriptor: | Outer membrane usher protein papC | | Authors: | Ford, B, Rego, A, Ragan, T.J, Pinkner, J, Dodson, K, Driscoll, P.C, Hultgren, S, Waksman, G. | | Deposit date: | 2010-01-19 | | Release date: | 2010-04-21 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural Homology between the C-Terminal Domain of the PapC Usher and Its Plug.
J.Bacteriol., 192, 2010
|
|
4MIB
 
 | | Hepatitis C Virus polymerase NS5B genotype 1b (BK) in complex with Compound 48 (N-({(3S)-1-[6-tert-butyl-5-methoxy-8-(2-oxo-1,2-dihydropyridin-3-yl)quinolin-3-yl]pyrrolidin-3-yl}methyl)methanesulfonamide) | | Descriptor: | DIMETHYL SULFOXIDE, N-({(3S)-1-[6-tert-butyl-5-methoxy-8-(2-oxo-1,2-dihydropyridin-3-yl)quinolin-3-yl]pyrrolidin-3-yl}methyl)methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | Authors: | Harris, S.F, Villasenor, A.G. | | Deposit date: | 2013-08-30 | | Release date: | 2014-05-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of N-[4-[6-tert-butyl-5-methoxy-8-(6-methoxy-2-oxo-1H-pyridin-3-yl)-3-quinolyl]phenyl]methanesulfonamide (RG7109), a potent inhibitor of the hepatitis C virus NS5B polymerase.
J.Med.Chem., 57, 2014
|
|
4MIA
 
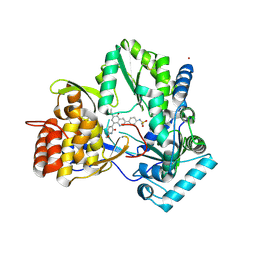 | | Hepatitis C Virus polymerase NS5B genotype 1b (BK) in complex with RG7109 (N-{4-[6-tert-butyl-5-methoxy-8-(6-methoxy-2-oxo-2,5-dihydropyridin-3-yl)quinolin-3-yl]phenyl}methanesulfonamide) | | Descriptor: | N-{4-[6-tert-butyl-5-methoxy-8-(6-methoxy-2-oxo-2,5-dihydropyridin-3-yl)quinolin-3-yl]phenyl}methanesulfonamide, RNA-directed RNA polymerase, ZINC ION | | Authors: | Harris, S.F, Villasenor, A.G. | | Deposit date: | 2013-08-30 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of N-[4-[6-tert-butyl-5-methoxy-8-(6-methoxy-2-oxo-1H-pyridin-3-yl)-3-quinolyl]phenyl]methanesulfonamide (RG7109), a potent inhibitor of the hepatitis C virus NS5B polymerase.
J.Med.Chem., 57, 2014
|
|
3EL8
 
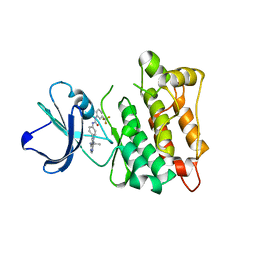 | | Crystal structure of c-Src in complex with pyrazolopyrimidine 5 | | Descriptor: | 1-{4-[4-amino-1-(1-methylethyl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl]phenyl}-3-[3-(trifluoromethyl)phenyl]urea, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Dar, A.C, Lopez, M.S, Shokat, K.M. | | Deposit date: | 2008-09-20 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small molecule recognition of c-Src via the Imatinib-binding conformation.
Chem.Biol., 15, 2008
|
|
3G01
 
 | |
3LG8
 
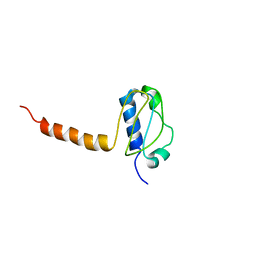 | | Crystal structure of the C-terminal part of subunit E (E101-206) from Methanocaldococcus jannaschii of A1AO ATP synthase | | Descriptor: | A-type ATP synthase subunit E | | Authors: | Balakrishna, A.M, Manimekalai, M.S.S, Hunke, C, Gayen, S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-01-19 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal and solution structure of the C-terminal part of the Methanocaldococcus jannaschii A1AO ATP synthase subunit E revealed by X-ray diffraction and small-angle X-ray scattering
J.Bioenerg.Biomembr., 42, 2010
|
|
7LRG
 
 | |
4AEX
 
 | |
2XHW
 
 | | HCV-J4 NS5B Polymerase Trigonal Crystal Form | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
3EL7
 
 | | Crystal structure of c-Src in complex with pyrazolopyrimidine 3 | | Descriptor: | 1-{3-[(4-amino-1-cyclopentyl-1H-pyrazolo[3,4-d]pyrimidin-3-yl)methyl]phenyl}-3-[3-(trifluoromethyl)phenyl]urea, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Dar, A.C, Lopez, M.S, Shokat, K.M. | | Deposit date: | 2008-09-20 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Small molecule recognition of c-Src via the Imatinib-binding conformation.
Chem.Biol., 15, 2008
|
|
1A7H
 
 | | GAMMA S CRYSTALLIN C-TERMINAL DOMAIN | | Descriptor: | GAMMAS CRYSTALLIN | | Authors: | Basak, A.K, Slingsby, C. | | Deposit date: | 1998-03-13 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The C-terminal domains of gammaS-crystallin pair about a distorted twofold axis.
Protein Eng., 11, 1998
|
|
3FZZ
 
 | | Structure of GrC | | Descriptor: | Granzyme C, SULFATE ION | | Authors: | Buckle, A.M, Kaiserman, D, Whisstock, J.C. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of granzyme C reveals an unusual mechanism of protease autoinhibition
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7JNT
 
 | |
3TEB
 
 | | endonuclease/exonuclease/phosphatase family protein from Leptotrichia buccalis C-1013-b | | Descriptor: | Endonuclease/exonuclease/phosphatase, MAGNESIUM ION | | Authors: | Chang, C, Bigelow, L, Muniez, I, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-12 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of endonuclease/exonuclease/phosphatase family protein from Leptotrichia buccalis C-1013-b
To be Published
|
|
4OO9
 
 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator mavoglurant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mavoglurant, Metabotropic glutamate receptor 5, ... | | Authors: | Dore, A.S, Okrasa, K, Patel, J.C, Serrano-Vega, M, Bennett, K, Cooke, R.M, Errey, J.C, Jazayeri, A, Khan, S, Tehan, B, Weir, M, Wiggin, G.R, Marshall, F.H. | | Deposit date: | 2014-01-31 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain.
Nature, 511, 2014
|
|
4OAY
 
 | | BldD CTD-c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | Authors: | Schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R.G. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
3N3T
 
 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase complex with cyclic di-gmp | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases.
J.Mol.Biol., 402, 2010
|
|
7Y3A
 
 | | Crystal structure of TRIM7 bound to 2C | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-2C | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3KNX
 
 | | HCV NS3 protease domain with P1-P3 macrocyclic ketoamide inhibitor | | Descriptor: | (2R)-2-{(3S,13S,16aS,17aR,17bS)-13-[({(1S)-1-[(4,4-dimethyl-2,6-dioxopiperidin-1-yl)methyl]-2,2-dimethylpropyl}carbamoyl)amino]-17,17-dimethyl-1,14-dioxooctadecahydro-2H-cyclopropa[3,4]pyrrolo[1,2-a][1,4]diazacyclohexadecin-3-yl}-2-hydroxy-N-prop-2-en-1-ylethanamide, BETA-MERCAPTOETHANOL, HCV NS3 Protease, ... | | Authors: | Venkatraman, S, Velazquez, F, Wu, W, Blackman, M, Chen, K.X, Bogen, S, Nair, L, Tong, X, Chase, R, Hart, A, Agrawal, S, Pichardo, J, Prongay, A, Cheng, K.-C, Girijavallabhan, V, Piwinski, J, Shih, N.-Y, Njoroge, F.G. | | Deposit date: | 2009-11-12 | | Release date: | 2010-10-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and structure-activity relationship of P1-P3 ketoamide derived macrocyclic inhibitors of hepatitis C virus NS3 protease.
J.Med.Chem., 52, 2009
|
|
