6LEM
 
 | | Structure of E. coli beta-glucuronidase complex with C6-nonyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-2-nonyl-4,5-bis(oxidanyl)piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Lin, H.-Y, Kuo, Y.-H, Lin, C.-H. | | Deposit date: | 2019-11-25 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LCP
 
 | | Cryo-EM structure of Dnf1 from Chaetomium thermophilum in the E2P state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, Y, Xu, J, Wu, X, Li, L. | | Deposit date: | 2019-11-19 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structures of a P4-ATPase lipid flippase in lipid bilayers.
Protein Cell, 11, 2020
|
|
6LFT
 
 | | Poa1p S30A mutant in complex with ADP-ribose | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
8CSG
 
 | | Human PRMT5:MEP50 structure with Fragment 1 and MTA Bound | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, 6-bromo-1H-pyrrolo[3,2-b]pyridin-5-amine, ... | | Authors: | Gunn, R.J, Lawson, J.D, Smith, C.R. | | Deposit date: | 2022-05-12 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
6LEG
 
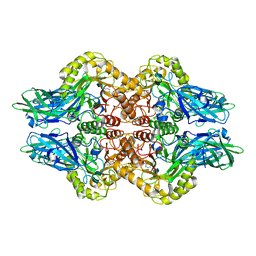 | | Structure of E. coli beta-glucuronidase complex with uronic isofagomine | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Lin, H.-Y, Kuo, Y.-H, Lin, C.-H. | | Deposit date: | 2019-11-25 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LFS
 
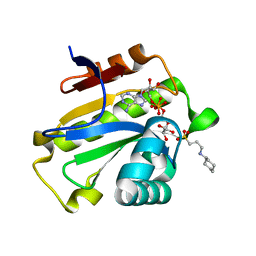 | | Poa1p H23A mutant in complex with ADP-ribose | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFZ
 
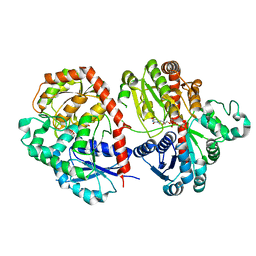 | | Crystal structure of SbCGTb in complex with UDPG | | Descriptor: | SbCGTb, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Gao, H.M, Yun, C.H. | | Deposit date: | 2019-12-04 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.866 Å) | | Cite: | Dissection of the general two-step di- C -glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7ZNK
 
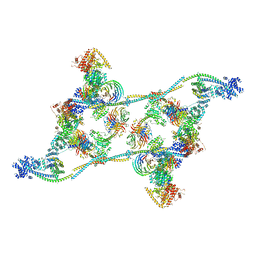 | | Structure of an endogenous human TREX complex bound to mRNA | | Descriptor: | RNA, Spliceosome RNA helicase DDX39B, THO complex subunit 1, ... | | Authors: | Pacheco-Fiallos, F.B, Vorlaender, M.K, Plaschka, C. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | mRNA recognition and packaging by the human transcription-export complex.
Nature, 616, 2023
|
|
8DCS
 
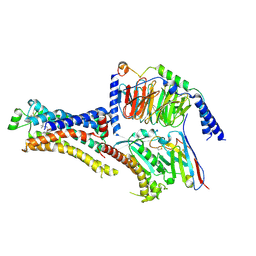 | | Cryo-EM structure of cyanopindolol-bound beta1-adrenergic receptor in complex with heterotrimeric Gs-protein | | Descriptor: | 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile, Endolysin,Endolysin,Beta-1 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Su, M, Paknejad, N, Hite, R.K, Huang, X.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structures of beta 1 -adrenergic receptor in complex with Gs and ligands of different efficacies.
Nat Commun, 13, 2022
|
|
7ZNL
 
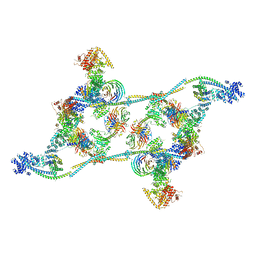 | | Structure of the human TREX core THO-UAP56 complex | | Descriptor: | Spliceosome RNA helicase DDX39B, THO complex subunit 1, THO complex subunit 2, ... | | Authors: | Pacheco-Fiallos, F.B, Vorlaender, M.K, Plaschka, C. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | mRNA recognition and packaging by the human transcription-export complex.
Nature, 616, 2023
|
|
6L1X
 
 | | Quinol-dependent nitric oxide reductase (qNOR) from Neisseria meningitidis in the monomeric oxidized state with zinc complex. | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | Authors: | Jamali, M.M.A, Antonyuk, S.V, Tosha, T, Muramoto, K, Hasnain, S.S, Shiro, Y. | | Deposit date: | 2019-10-01 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The active form of quinol-dependent nitric oxide reductase fromNeisseria meningitidisis a dimer.
Iucrj, 7, 2020
|
|
4WMS
 
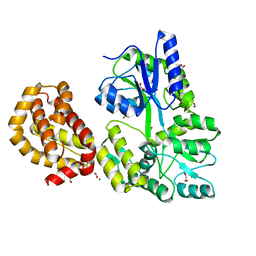 | | STRUCTURE OF APO MBP-MCL1 AT 1.9A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
6L6N
 
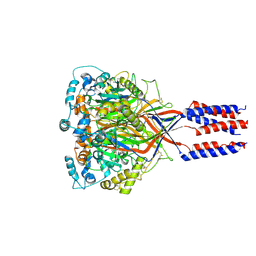 | | hASIC1a co-crystallized with Nafamostat | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lei, F, Jian, S. | | Deposit date: | 2019-10-29 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | hASIC1a co-crystallized with Mamb-1
To Be Published
|
|
5YLK
 
 | | Complex structure of GH 113 family beta-1,4-mannanase with mannobiose | | Descriptor: | beta-1,4-mannanase, beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase fromn Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
6L47
 
 | | Structure of the human sterol O-acyltransferase 1 in complex with CI-976 | | Descriptor: | 2,2-dimethyl-N-(2,4,6-trimethoxyphenyl)dodecanamide, CHOLESTEROL, Sterol O-acyltransferase 1 | | Authors: | Chen, L, Guan, C, Niu, Y. | | Deposit date: | 2019-10-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor.
Nat Commun, 11, 2020
|
|
4WT8
 
 | | Crystal Structure of bactobolin A bound to 70S ribosome-tRNA complex | | Descriptor: | 23S rRNA (2899-MER), 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Amunts, A, Fiedorczuk, K, Ramakrishnan, V. | | Deposit date: | 2014-10-29 | | Release date: | 2015-01-21 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bactobolin A Binds to a Site on the 70S Ribosome Distinct from Previously Seen Antibiotics.
J.Mol.Biol., 427, 2015
|
|
6L90
 
 | | Crystal structure of ugt transferase enzyme | | Descriptor: | Glycosyltransferase, SULFATE ION | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2019-11-07 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 2020
|
|
4WR4
 
 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7bGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
8DCR
 
 | | Cryo-EM structure of dobutamine-bound beta1-adrenergic receptor in complex with heterotrimeric Gs-protein | | Descriptor: | DOBUTAMINE, Endolysin,Endolysin,Beta-1 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Su, M, Paknejad, N, Hite, R.K, Huang, X.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of beta 1 -adrenergic receptor in complex with Gs and ligands of different efficacies.
Nat Commun, 13, 2022
|
|
4WZ9
 
 | | APN1 from Anopheles gambiae | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, AGAP004809-PA, ALA-ALA-ALA-LYS-ALA, ... | | Authors: | Atkinson, S.C, Armistead, J.S, Mathias, D.K, Sandeu, M.M, Tao, D, Borhani-Dizaji, N, Morlais, I, Dinglasan, R.R, Borg, N.A. | | Deposit date: | 2014-11-19 | | Release date: | 2015-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Anopheles-midgut APN1 structure reveals a new malaria transmission-blocking vaccine epitope.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6LI0
 
 | | Crystal structure of GPR52 in complex with agonist c17 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRATE ANION, Chimera of G-protein coupled receptor 52 and Flavodoxin, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LIG
 
 | |
7ZQT
 
 | |
7QTT
 
 | |
6LFQ
 
 | | Crystal structure of Poa1p in apo form | | Descriptor: | ADP-ribose 1''-phosphate phosphatase, GLYCEROL | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
