6MVK
 
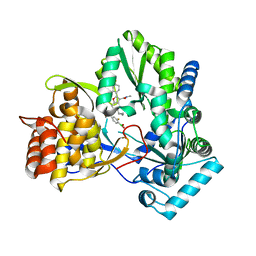 | | HCV NS5B 1b N316 bound to Compound 18 | | Descriptor: | (4-{(4S)-3-[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl]-2-oxo-1,3-oxazolidin-4-yl}-2-fluorophenyl)boronic acid, HCV Polymerase | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
5C83
 
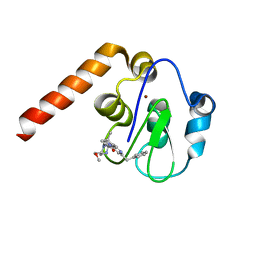 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 21 | | Descriptor: | (2R,5R)-4-[2-(6-benzyl-3,3-dimethyl-2,3-dihydro-1H-pyrrolo[3,2-c]pyridin-1-yl)-2-oxoethyl]-5-(methoxymethyl)-2-methylpiperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-25 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
4YMJ
 
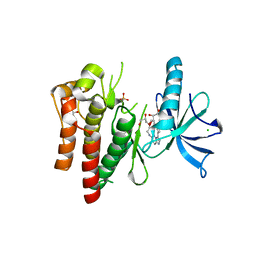 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 4-[6-(benzylamino)imidazo[1,2-b]pyridazin-3-yl]benzonitrile, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-06 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
6VMZ
 
 | | Crystal Structure of a H5N1 influenza virus hemagglutinin with CBS1117 | | Descriptor: | 2,6-dichloro-N-[1-(propan-2-yl)piperidin-4-yl]benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Antanasijevic, A, Durst, M.A, Lavie, A, Caffrey, M. | | Deposit date: | 2020-01-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of avian influenza hemagglutinin in complex with a small molecule entry inhibitor.
Life Sci Alliance, 3, 2020
|
|
5V7Q
 
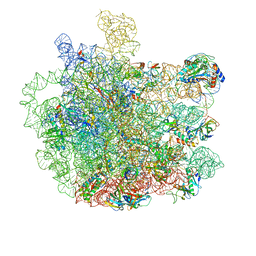 | | Cryo-EM structure of the large ribosomal subunit from Mycobacterium tuberculosis bound with a potent linezolid analog | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Yang, K, Chang, J.-Y, Cui, Z, Zhang, J. | | Deposit date: | 2017-03-20 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis.
Nucleic Acids Res., 45, 2017
|
|
5UNG
 
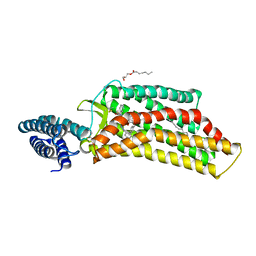 | | XFEL structure of human angiotensin II type 2 receptor (Orthorhombic form) in complex with compound 1 (N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl] methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera protein of Type-2 angiotensin II receptor and Soluble cytochrome b562, N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide, ... | | Authors: | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | Deposit date: | 2017-01-30 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
5XLY
 
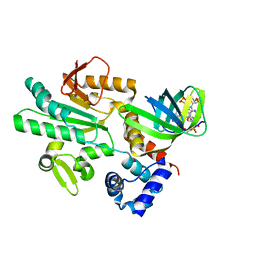 | | Crystal structure of CheR1 in complex with c-di-GMP-bound MapZ | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Chemotaxis protein methyltransferase 1, Cyclic diguanosine monophosphate-binding protein PA4608 | | Authors: | Yuan, Z, Zhu, Y, Gu, L. | | Deposit date: | 2017-05-12 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Structural basis for the regulation of chemotaxis by MapZ in the presence of c-di-GMP
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5X5G
 
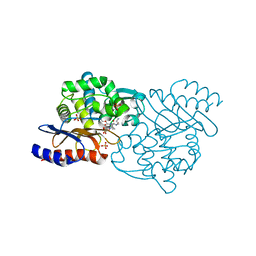 | | Crystal structure of TLA-3 extended-spectrum beta-lactamase in a complex with OP0595 | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, SODIUM ION, ... | | Authors: | Wachino, J, Jin, W, Arakawa, Y. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Antimicrob. Agents Chemother., 61, 2017
|
|
5SQT
 
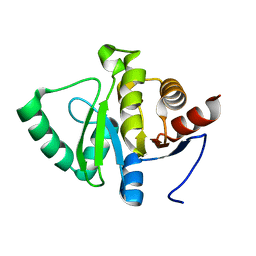 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000833624464 - (R,R) and (S,S) isomers | | Descriptor: | (3R,4R)-4-methyl-1-(2-oxo-2,3-dihydro-1,3-benzoxazole-7-carbonyl)pyrrolidine-3-carboxylic acid, (3S,4S)-4-methyl-1-(2-oxo-2,3-dihydro-1,3-benzoxazole-7-carbonyl)pyrrolidine-3-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2CFA
 
 | | Structure of viral flavin-dependant thymidylate synthase ThyX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, THYMIDYLATE SYNTHASE | | Authors: | Graziani, S, Bernauer, J, Skouloubris, S, Graille, M, Zhou, C.-Z, Marchand, C, Decottignies, P, van Tilbeurgh, H, Myllykallio, H, Liebl, U. | | Deposit date: | 2006-02-17 | | Release date: | 2006-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic Mechanism and Structure of Viral Flavin-Dependent Thymidylate Synthase Thyx.
J.Biol.Chem., 281, 2006
|
|
6N5G
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 2 | | Descriptor: | 4-[(quinolin-3-yl)methyl]-N-[4-(trifluoromethoxy)phenyl]piperidine-1-carboxamide, Epoxide hydrolase TrEH | | Authors: | Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
4CYP
 
 | | Leishmania major N-myristoyltransferase in complex with a pyrrolidine inhibitor. | | Descriptor: | (3R)-4-(4-chlorophenyl)-1-[(3S,4R)-3-(4-chlorophenyl)-4-(hydroxymethyl)pyrrolidin-1-yl]-3-hydroxybutan-1-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Hutton, J.A, Goncalves, V, Brannigan, J.A, Paape, D, Waugh, T, Roberts, S.M, Bell, A.S, Wilkinson, A.J, Smith, D.F, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2014-04-14 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Based Design of Potent and Selective Leishmania N- Myristoyltransferase Inhibitors.
J.Med.Chem., 57, 2014
|
|
2YJ2
 
 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | Descriptor: | (2S,4R)-1-[1-(4-BROMOPHENYL)CYCLOPROPYL]CARBONYL-4-(2-CHLOROPHENYL)SULFONYL-N-[1-(IMINOMETHYL)CYCLOPROPYL]PYRROLIDINE-2-CARBOXAMIDE, CATHEPSIN L1, GLYCEROL | | Authors: | Banner, D.W, Benz, J.M, Haap, W. | | Deposit date: | 2011-05-18 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Halogen Bonding at the Active Sites of Human Cathepsin L and Mek1 Kinase: Efficient Interactions in Different Environments.
Chemmedchem, 6, 2011
|
|
4TW6
 
 | | The Fk1 domain of FKBP51 in complex with iFit1 | | Descriptor: | (3-{(1R)-3-(3,4-dimethoxyphenyl)-1-[({(2S)-1-[(2S)-2-(3,4,5-trimethoxyphenyl)pent-4-enoyl]piperidin-2-yl}carbonyl)oxy]propyl}phenoxy)acetic acid, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | Authors: | Gaali, S, Kirschner, A, Cuboni, S, Hartmann, J, Kozany, C, Balsevich, G, Namendorf, C, Fernandez-Vizarra, P, Almeida, O.F.X, Ruehter, G, Uhr, M, Schmidt, M.V, Touma, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective inhibitors of the FK506-binding protein 51 by induced fit.
Nat.Chem.Biol., 11, 2015
|
|
4TW8
 
 | | The Fk1-Fk2 domains of FKBP52 in complex with iFit-FL | | Descriptor: | 2-(5-{[({3-[(1R)-1-[({(2S)-1-[(2S)-2-[(1S)-cyclohex-2-en-1-yl]-2-(3,4,5-trimethoxyphenyl)acetyl]piperidin-2-yl}carbonyl)oxy]-3-(3,4-dimethoxyphenyl)propyl]phenoxy}acetyl)amino]methyl}-6-hydroxy-3-oxo-3H-xanthen-9-yl)benzoic acid, Peptidyl-prolyl cis-trans isomerase FKBP4 | | Authors: | Gaali, S, Kirschner, A, Cuboni, S, Hartmann, J, Kozany, C, Balsevich, G, Namendorf, C, Fernandez-Vizarra, P, Almeida, O.F.X, Ruehter, G, Uhr, M, Schmidt, M.V, Touma, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Selective inhibitors of the FK506-binding protein 51 by induced fit.
Nat.Chem.Biol., 11, 2015
|
|
7GM7
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-a577c8a2-1 (Mpro-P2099) | | Descriptor: | (4S)-6-chloro-N-(isoquinolin-4-yl)-1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
3KPZ
 
 | | Crystal structure of a novel vitamin D3 analogue, ZK203278 showing dissociated profile | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-{(1R,3aS,7aR)-1-[(1R,5S)-5-hydroxy-1-methyl-5-(1,3-thiazol-2-yl)pentyl]-7a-methyloctahydro-4H-inden-4-ylidene}ethylidene]-4-methylidenecyclohexane-1,3-diol, Vitamin D3 receptor | | Authors: | Rochel, N, Moras, D. | | Deposit date: | 2009-11-17 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a vitamin D3 analog, ZK203278, showing dissociated profile.
Anticancer Res., 32, 2012
|
|
7GNI
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-b1ef7fe3-1 (Mpro-P2660) | | Descriptor: | 2-[(3'S)-6-chloro-1'-(6-chloroisoquinolin-4-yl)-1,2'-dioxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylacetamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GNU
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-133e7cd9-2 (Mpro-P3074) | | Descriptor: | 1-{[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-2'-oxo-1H-spiro[isoquinoline-4,3'-piperidine]-2(3H)-sulfonyl]methyl}cyclopropane-1-carbonitrile, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6VLM
 
 | |
5AE1
 
 | | Ether Lipid-Generating Enzyme AGPS in complex with inhibitor ZINC69435460 | | Descriptor: | (3-(2-FLUOROPHENYL)-N-(1-(2-OXO-2,3-DIHYDRO-1H-BENZO[D]IMIDAZOL-5-YL)ETHYL)BUTANAMIDE), ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, PEROXISOMAL, ... | | Authors: | Piano, V, Benjamin, D.I, Valente, S, Nenci, S, Marrocco, B, Mai, A, Aliverti, A, Nomura, D.K, Mattevi, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Inhibitors for the Ether Lipid-Generating Enzyme Agps as Anti-Cancer Agents.
Acs Chem.Biol., 10, 2015
|
|
6VRB
 
 | | Cryo-EM structure of AcrVIA1-Cas13(crRNA) complex | | Descriptor: | AcrVIA1, CRISPR-associated endoribonuclease Cas13a, RNA (52-MER) | | Authors: | Jia, N, Meeske, A.J, Marraffini, L.A, Patel, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity.
Science, 369, 2020
|
|
4TWC
 
 | | 2-Benzamido-N-(1H-benzo[d]imidazol-2-yl)thiazole-4- carboxamide derivatives as potent inhibitors of CK1d/e | | Descriptor: | 2-{[2-(trifluoromethoxy)benzoyl]amino}-N-[6-(trifluoromethyl)-1H-benzimidazol-2-yl]-1,3-thiazole-4-carboxamide, Casein kinase I isoform delta, DIMETHYL SULFOXIDE, ... | | Authors: | Bischof, J, Leban, L, Zaja, M, Grothey, A, Radunsky, B, Othersen, O, Strobl, S, Vitt, D, Knippschild, U. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Benzamido-N-(1H-benzo[d]imidazol-2-yl)thiazole-4-carboxamide derivatives as potent inhibitors of CK1d/e.
Amino Acids, 43, 2012
|
|
4U15
 
 | | M3-mT4L receptor bound to tiotropium | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, D(-)-TARTARIC ACID, ... | | Authors: | Thorsen, T.S, Matt, R, Weis, W.I, Kobilka, B. | | Deposit date: | 2014-07-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis.
Structure, 22, 2014
|
|
5AAB
 
 | | Structure of C1156Y,L1198F Mutant Human Anaplastic Lymphoma Kinase in Complex with Crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Resensitization to Crizotinib by the Lorlatinib Alk Resistance Mutation L1198F.
N.Engl.J.Med., 374, 2016
|
|
