7U6M
 
 | |
5TMP
 
 | |
1XYA
 
 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | HYDROXIDE ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
1XYC
 
 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | 3-O-METHYLFRUCTOSE IN LINEAR FORM, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
1XYB
 
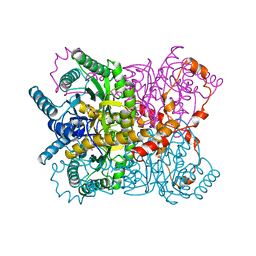 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | D-glucose, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
4TMK
 
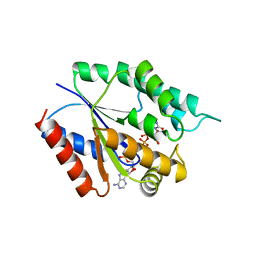 | |
3TMK
 
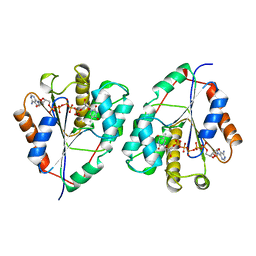 | | CRYSTAL STRUCTURE OF YEAST THYMIDYLATE KINASE COMPLEXED WITH THE BISUBSTRATE INHIBITOR TP5A AT 2.0 A RESOLUTION: IMPLICATIONS FOR CATALYSIS AND AZT ACTIVATION | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, THYMIDYLATE KINASE | | Authors: | Lavie, A, Schlichting, I, Konrad, M, Goody, R.S, Brundiers, R, Reinstein, J. | | Deposit date: | 1998-01-26 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of yeast thymidylate kinase complexed with the bisubstrate inhibitor P1-(5'-adenosyl) P5-(5'-thymidyl) pentaphosphate (TP5A) at 2.0 A resolution: implications for catalysis and AZT activation.
Biochemistry, 37, 1998
|
|
1TMK
 
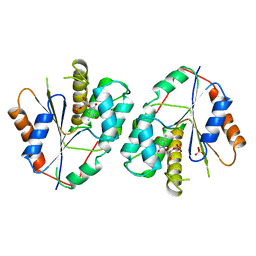 | |
3QEO
 
 | | S74E-R104M-D133A dCK variant in complex with L-deoxythymidine and UDP | | Descriptor: | Deoxycytidine kinase, L-deoxythymidine, URIDINE-5'-DIPHOSPHATE | | Authors: | Lavie, A, Hazra, S. | | Deposit date: | 2011-01-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Post-translational phosphorylation of serine 74 of human deoxycytidine kinase favors the enzyme adopting the open conformation making it competent for nucleoside binding and release.
Biochemistry, 50, 2011
|
|
3QEN
 
 | | S74E dCK in complex with 5-bromodeoxycytidine and UDP | | Descriptor: | 5-bromo-2'-deoxycytidine, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Lavie, A, Hazra, S. | | Deposit date: | 2011-01-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Post-translational phosphorylation of serine 74 of human deoxycytidine kinase favors the enzyme adopting the open conformation making it competent for nucleoside binding and release.
Biochemistry, 50, 2011
|
|
3MTR
 
 | | Crystal structure of the Ig5-FN1 tandem of human NCAM | | Descriptor: | Neural cell adhesion molecule 1, SULFATE ION | | Authors: | Lavie, A, Foley, D.A. | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mutagenesis of neural cell adhesion molecule domains: evidence for flexibility in the placement of polysialic acid attachment sites
J.Biol.Chem., 285, 2010
|
|
2TMK
 
 | |
2RCZ
 
 | | Structure of the second PDZ domain of ZO-1 | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Lavie, A, Lye, M.F. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Domain swapping within PDZ2 is responsible for dimerization of ZO proteins.
J.Biol.Chem., 282, 2007
|
|
2GYI
 
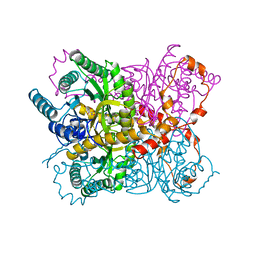 | | DESIGN, SYNTHESIS, AND CHARACTERIZATION OF A POTENT XYLOSE ISOMERASE INHIBITOR, D-THREONOHYDROXAMIC ACID, AND HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHIC STRUCTURE OF THE ENZYME-INHIBITOR COMPLEX | | Descriptor: | 2,3,4,N-TETRAHYDROXY-BUTYRIMIDIC ACID, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-09-01 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, Synthesis, and Characterization of a Potent Xylose Isomerase Inhibitor, D-Threonohydroxamic Acid, and High-Resolution X-Ray Crystallographic Structure of the Enzyme-Inhibitor Complex
Biochemistry, 34, 1995
|
|
4YYX
 
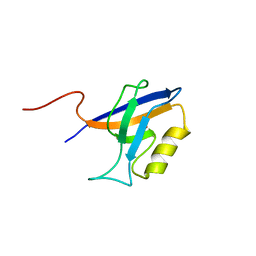 | |
6VMZ
 
 | | Crystal Structure of a H5N1 influenza virus hemagglutinin with CBS1117 | | Descriptor: | 2,6-dichloro-N-[1-(propan-2-yl)piperidin-4-yl]benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Antanasijevic, A, Durst, M.A, Lavie, A, Caffrey, M. | | Deposit date: | 2020-01-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of avian influenza hemagglutinin in complex with a small molecule entry inhibitor.
Life Sci Alliance, 3, 2020
|
|
4ECD
 
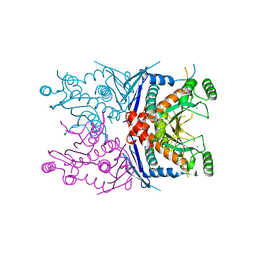 | | 2.5 Angstrom Resolution Crystal Structure of Bifidobacterium longum Chorismate Synthase | | Descriptor: | CHLORIDE ION, Chorismate synthase | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2.5 Angstrom Resolution Crystal Structure of Bifidobacterium longum Chorismate Synthase
TO BE PUBLISHED
|
|
5W6O
 
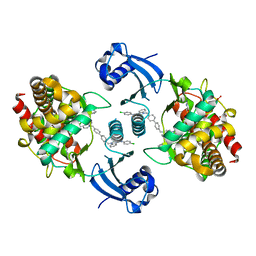 | | Choline Kinase Alpha in Complex with TCD-717 | | Descriptor: | 1,1'-[[1,1'-biphenyl]-4,4'-diylbis(methylene)]bis{4-[(4-chlorophenyl)(methyl)amino]quinolin-1-ium}, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kall, S.K, Lavie, A. | | Deposit date: | 2017-06-16 | | Release date: | 2018-02-14 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of a Unique Inhibitor-Binding Site on Choline Kinase alpha.
Biochemistry, 57, 2018
|
|
1LVG
 
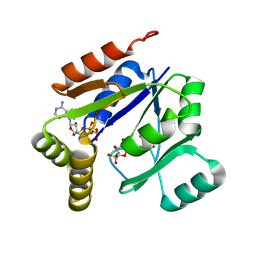 | | Crystal structure of mouse guanylate kinase in complex with GMP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Guanylate kinase, ... | | Authors: | Sekulic, N, Shuvalova, L, Spangenberg, O, Konrad, M, Lavie, A. | | Deposit date: | 2002-05-28 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the closed
conformation of mouse guanylate kinase.
J.Biol.Chem., 277, 2002
|
|
1KGD
 
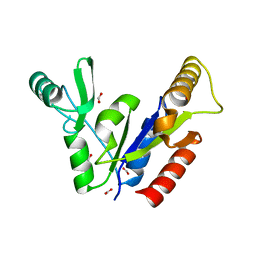 | | Crystal Structure of the Guanylate Kinase-like Domain of Human CASK | | Descriptor: | FORMIC ACID, PERIPHERAL PLASMA MEMBRANE CASK | | Authors: | Li, Y, Spangenberg, O, Paarmann, I, Konrad, M, Lavie, A. | | Deposit date: | 2001-11-26 | | Release date: | 2001-12-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.314 Å) | | Cite: | Structural basis for nucleotide-dependent regulation of membrane-associated guanylate kinase-like domains.
J.Biol.Chem., 277, 2002
|
|
5K28
 
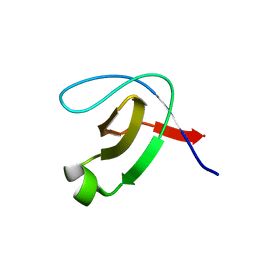 | | Structure of the unbound SH3 domain of MLK3 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 11 | | Authors: | Kall, S.K, Lavie, A. | | Deposit date: | 2016-05-18 | | Release date: | 2017-12-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of two distinct peptide-binding pockets in the SH3 domain of human mixed-lineage kinase 3.
J. Biol. Chem., 293, 2018
|
|
5K26
 
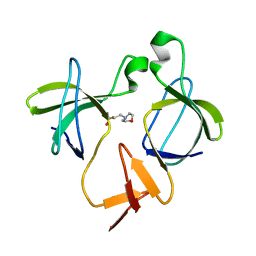 | |
2HAZ
 
 | | Crystal structure of the first fibronectin domain of human NCAM1 | | Descriptor: | Neural cell adhesion molecule 1, SODIUM ION | | Authors: | Sekulic, N, Lavie, A. | | Deposit date: | 2006-06-13 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel alpha-helix in the first fibronectin type III repeat of the neural cell adhesion molecule is critical for N-glycan polysialylation.
J.Biol.Chem., 281, 2006
|
|
6PCX
 
 | | Crystal Structure of a H5N1 influenza virus hemagglutinin at pH 6.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hemagglutinin, ... | | Authors: | Antanasijevic, A, Durst, M.A, Lavie, A, Caffrey, M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification of a pH sensor in Influenza hemagglutinin using X-ray crystallography.
J.Struct.Biol., 209, 2020
|
|
6PD6
 
 | | Crystal Structure of a H5N1 influenza virus hemagglutinin at pH 7.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hemagglutinin, ... | | Authors: | Antanasijevic, A, Durst, M.A, Lavie, A, Caffrey, M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Identification of a pH sensor in Influenza hemagglutinin using X-ray crystallography.
J.Struct.Biol., 209, 2020
|
|
