7ZMK
 
 | |
5TR8
 
 | | Crystal structure of vaccine-elicited pan- influenza H1N1 neutralizing murine antibody 441D6. | | Descriptor: | 441D6 Fab Heavy chain, 441D6 Fab Light chain, NICKEL (II) ION | | Authors: | Joyce, M.G, Kanekiyo, M, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2016-10-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses.
Nat.Immunol., 20, 2019
|
|
5T5N
 
 | | Calcium-activated chloride channel bestrophin-1 (BEST1), triple mutant: I76A, F80A, F84A; in complex with an Fab antibody fragment, chloride, and calcium | | Descriptor: | CALCIUM ION, CHLORIDE ION, Fab antibody fragment, ... | | Authors: | Long, S.B, Vaisey, G. | | Deposit date: | 2016-08-31 | | Release date: | 2016-11-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct regions that control ion selectivity and calcium-dependent activation in the bestrophin ion channel.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6RPS
 
 | | X-ray crystal structure of carbonic anhydrase XII complexed with a theranostic monoclonal antibody fragment | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Alterio, V, Esposito, D, De Simone, G. | | Deposit date: | 2019-05-14 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Biochemical and Structural Insights into Carbonic Anhydrase XII/Fab6A10 Complex.
J.Mol.Biol., 431, 2019
|
|
8JNK
 
 | |
8JNR
 
 | |
7CU5
 
 | | N-Glycosylation of PD-1 and glycosylation dependent binding of PD-1 specific monoclonal antibody camrelizumab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Programmed cell death protein 1, ... | | Authors: | Liu, K.F, Tan, S.G, Jin, W.J, Guan, J.W, Wang, W.L, Sun, H, Qi, J.X, Yan, J.H, Chai, Y, Wang, Z.F, Chu, X.D, Gao, G.F. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | N-glycosylation of PD-1 promotes binding of camrelizumab.
Embo Rep., 21, 2020
|
|
1F58
 
 | | IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 24-RESIDUE PEPTIDE (RESIDUES 308-333 OF HIV-1 GP120 (MN ISOLATE) WITH ALA TO AIB SUBSTITUTION AT POSITION 323 | | Descriptor: | Envelope glycoprotein gp120, PROTEIN (IGG1 ANTIBODY 58.2 (HEAVY CHAIN)), PROTEIN (IGG1 ANTIBODY 58.2 (LIGHT CHAIN)) | | Authors: | Stanfield, R.L, Cabezas, E, Satterthwait, A.C, Stura, E.A, Profy, A.T, Wilson, I.A. | | Deposit date: | 1998-10-21 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs.
Structure Fold.Des., 7, 1999
|
|
6Q1G
 
 | |
6WOQ
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 1a bound to neutralizing antibody HC1AM and non neutralizing antibody E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.667 Å) | | Cite: | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
6WO5
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 1a bound to neutralizing antibody 212.1.1 and non neutralizing antibody E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2020-04-24 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
6NZ7
 
 | |
4EIZ
 
 | | Structure of Nb113 bound to apoDHFR | | Descriptor: | Dihydrofolate reductase, Nb113 Camel antibody fragment | | Authors: | Oyen, D, Srinivasan, V. | | Deposit date: | 2012-04-06 | | Release date: | 2013-04-24 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihyrofolate reductase of Escherichia coli.
Biochim.Biophys.Acta, 1834, 2013
|
|
7Y58
 
 | |
7A3R
 
 | | Crystal structure of dengue 1 virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Core protein, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
6LNT
 
 | | Cryo-EM structure of immature Zika virus in complex with human antibody DV62.5 Fab | | Descriptor: | Envelope protein, Fab DV62.5 heavy-chain variable region, Fab DV62.5 light-chain variable region, ... | | Authors: | Tan, T.Y, Fibriansah, G, Kostyuchenko, V.A, Ng, T.S, Lim, X.X, Lim, X.N, Shi, J, Morais, M.C, Corti, D, Lok, S.M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Capsid protein structure in Zika virus reveals the flavivirus assembly process.
Nat Commun, 11, 2020
|
|
2UZI
 
 | | Crystal structure of HRAS(G12V) - anti-RAS Fv complex | | Descriptor: | ANTI-RAS FV HEAVY CHAIN, ANTI-RAS FV LIGHT CHAIN, GTPASE HRAS, ... | | Authors: | Tanaka, T, williams, R.L, Rabbitts, T.H. | | Deposit date: | 2007-04-27 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tumour Prevention by a Single Antibody Domain Targeting the Interaction of Signal Transduction Proteins with Ras.
Embo J., 26, 2007
|
|
8FXB
 
 | | SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
8FXC
 
 | | SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
6CW3
 
 | | Crystal structure of a yeast SAGA transcriptional coactivator Ada2/Gcn5 HAT subcomplex, crystal form 2 | | Descriptor: | Histone acetyltransferase GCN5, Transcriptional adapter 2, ZINC ION, ... | | Authors: | Sun, J, Paduch, M, Kim, S.A, Kramer, R.M, Barrios, A.F, Lu, V, Luke, J, Usatyuk, S, Kossiakoff, A.A, Tan, S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-19 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CW2
 
 | | Crystal structure of a yeast SAGA transcriptional coactivator Ada2/Gcn5 HAT subcomplex, crystal form 1 | | Descriptor: | Histone acetyltransferase GCN5, Transcriptional adapter 2, ZINC ION, ... | | Authors: | Sun, J, Paduch, M, Kim, S.A, Kramer, R.M, Barrios, A.F, Lu, V, Luke, J, Usatyuk, S, Kossiakoff, A.A, Tan, S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6I1O
 
 | | Fab fragment of an antibody selective for wild-type alpha-1-antitrypsin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FAB 2H2 heavy chain, FAB 2H2 light chain, ... | | Authors: | Laffranchi, M, Elliston, E.L.K, Miranda, E, Perez, J, Fra, A, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-10-29 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Intrahepatic heteropolymerization of M and Z alpha-1-antitrypsin.
JCI Insight, 5, 2020
|
|
6I3Z
 
 | | Fab fragment of an antibody selective for wild-type alpha-1-antitrypsin in complex with its antigen | | Descriptor: | Alpha-1-antitrypsin, Fab 2H2 heavy chain, Fab 2H2 light chain, ... | | Authors: | Laffranchi, M, Elliston, E.L.K, Miranda, E, Perez, J, Jagger, A.M, Fra, A, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-11-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Intrahepatic heteropolymerization of M and Z alpha-1-antitrypsin.
JCI Insight, 5, 2020
|
|
4PB9
 
 | | Structure of the Fab fragment of the anti-Francisella tularensis GroEL antibody Ab64 | | Descriptor: | Ab64 heavy chain, Ab64 light chain, SULFATE ION | | Authors: | Lu, Z, Rynkiewicz, M.J, Seaton, B.A, Sharon, J. | | Deposit date: | 2014-04-11 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | B-Cell Epitopes in GroEL of Francisella tularensis.
Plos One, 9, 2014
|
|
4PB0
 
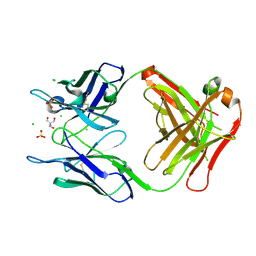 | | Structure of the Fab fragment of the anti-Francisella tularensis GroEL antibody Ab53 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ab53 heavy chain, Ab53 light chain, ... | | Authors: | Lu, Z, Rynkiewicz, M.J, Seaton, B.A, Sharon, J. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | B-Cell Epitopes in GroEL of Francisella tularensis.
Plos One, 9, 2014
|
|
