6XQZ
 
 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidoglycan D,D-transpeptidase PenA, ... | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6VOT
 
 | | Crystal structure of Pseudomonas aerugonisa PBP3 complexed to gamma-lactam YU253434 | | Descriptor: | 1-[(2S)-2-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-3-oxopropyl]-4-{[2-(5,6 -dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl]carbamoyl}-2,5-dihydro-1H-pyrazole-3-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F. | | Deposit date: | 2020-01-31 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A gamma-Lactam Siderophore Antibiotic Effective against Multidrug-Resistant Gram-Negative Bacilli.
J.Med.Chem., 63, 2020
|
|
6NTZ
 
 | | Crystal structure of E. coli PBP5-meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, D-alanyl-D-alanine carboxypeptidase | | Authors: | Caveney, N.A, Strynadka, N.C.J, Caballero, G, Worrall, L.J. | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insight into YcbB-mediated beta-lactam resistance in Escherichia coli.
Nat Commun, 10, 2019
|
|
4FSF
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 complexed with compound 14 | | Descriptor: | (4R,5S,8Z)-8-(2-amino-1,3-thiazol-4-yl)-1-[3-(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)-1,2-oxazol-5-yl]-5-formyl-11,11-dimethyl-1,7-dioxo-4-(sulfoamino)-10-oxa-2,6,9-triazadodec-8-en-12-oic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel monobactams utilizing a siderophore uptake mechanism for the treatment of gram-negative infections.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6UN3
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with ticarcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2019-10-10 | | Release date: | 2019-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6UN1
 
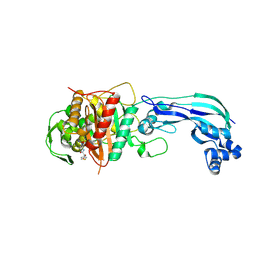 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with temocillin | | Descriptor: | (2R,4S)-2-[(1S)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-1-methoxy-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4 -carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2019-10-10 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6XQV
 
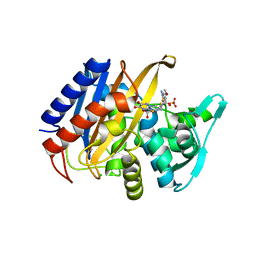 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae in a pre-acylation complex with ceftriaxone | | Descriptor: | CHLORIDE ION, Ceftriaxone, Probable peptidoglycan D,D-transpeptidase PenA, ... | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6XQY
 
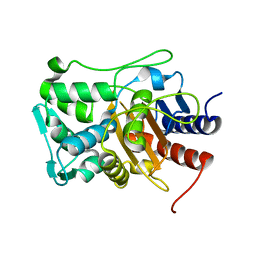 | |
6XQX
 
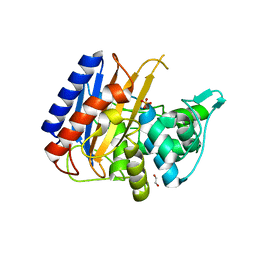 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae with the H514A mutation at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, Probable peptidoglycan D,D-transpeptidase PenA, SULFATE ION | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6BAS
 
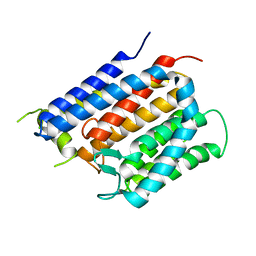 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA D255A mutant (Q5SIX3_THET8) | | Descriptor: | CHLORIDE ION, Peptidoglycan glycosyltransferase RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
7LC4
 
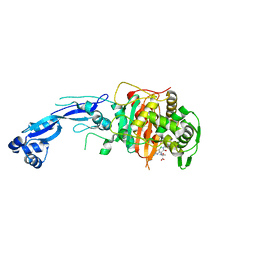 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with gamma-lactam YU253911 | | Descriptor: | 1-[(2S)-2-{[(2Z)-2-(2-amino-5-chloro-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-3-oxopropyl]-4-{[2-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl]carbamoyl}-2,5-dihydro-1H-pyrazole-3-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2021-01-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A gamma-lactam siderophore antibiotic effective against multidrug-resistant Pseudomonas aeruginosa, Klebsiella pneumoniae, and Acinetobacter spp.
Eur.J.Med.Chem., 220, 2021
|
|
1SKF
 
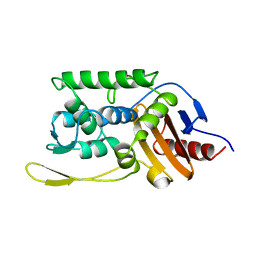 | | CRYSTAL STRUCTURE OF THE STREPTOMYCES K15 DD-TRANSPEPTIDASE | | Descriptor: | D-ALANYL-D-ALANINE TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 1998-08-20 | | Release date: | 1999-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a penicilloyl-serine transferase of intermediate penicillin sensitivity. The DD-transpeptidase of streptomyces K15.
J.Biol.Chem., 274, 1999
|
|
8BH1
 
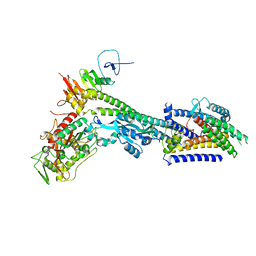 | | Core divisome complex FtsWIQBL from Pseudomonas aeruginosa | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ, ... | | Authors: | Kaeshammer, L, van den Ent, F, Jeffery, M, Lowe, J. | | Deposit date: | 2022-10-28 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the bacterial divisome core complex and antibiotic target FtsWIQBL.
Nat Microbiol, 8, 2023
|
|
4YE5
 
 | | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703 | | Descriptor: | ACETATE ION, GLYCEROL, Peptidoglycan synthetase penicillin-binding protein 3 | | Authors: | Cuff, M, Tan, K, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703
To Be Published
|
|
6O9S
 
 | |
6O9W
 
 | |
8DA2
 
 | | Acinetobacter baumannii L,D-transpeptidase | | Descriptor: | L,D-transpeptidase family protein | | Authors: | Toth, M, Stewart, N.K, Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2022-06-12 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The l,d-Transpeptidase Ldt Ab from Acinetobacter baumannii Is Poorly Inhibited by Carbapenems and Has a Unique Structural Architecture.
Acs Infect Dis., 8, 2022
|
|
1ZAT
 
 | | Crystal Structure of an Enterococcus faecium peptidoglycan binding protein at 2.4 A resolution | | Descriptor: | L,D-transpeptidase, SULFATE ION, ZINC ION | | Authors: | Biarrotte-Sorin, S, Hugonnet, J.-E, Mainardi, J.-L, Gutmann, L, Rice, L, Arthur, M, Mayer, C. | | Deposit date: | 2005-04-07 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a Novel beta-Lactam-insensitive Peptidoglycan Transpeptidase.
J.Mol.Biol., 359, 2006
|
|
1XA1
 
 | | Crystal structure of the sensor domain of BlaR1 from Staphylococcus aureus in its apo form | | Descriptor: | PHOSPHATE ION, PYROPHOSPHATE 2-, Regulatory protein blaR1 | | Authors: | Wilke, M.S, Hills, T.L, Zhang, H.Z, Chambers, H.F, Strynadka, N.C. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus.
J.Biol.Chem., 279, 2004
|
|
4DKI
 
 | | Structural Insights into the Anti- Methicillin-Resistant Staphylococcus aureus (MRSA) Activity of Ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, BICARBONATE ION, CADMIUM ION, ... | | Authors: | Lovering, A.L, Gretes, M.C, Strynadka, N.C.J. | | Deposit date: | 2012-02-03 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights into the Anti-methicillin-resistant Staphylococcus aureus (MRSA) Activity of Ceftobiprole.
J.Biol.Chem., 287, 2012
|
|
1XA7
 
 | | Crystal structure of the benzylpenicillin-acylated BlaR1 sensor domain from Staphylococcus aureus | | Descriptor: | OPEN FORM - PENICILLIN G, Regulatory protein BlaR1 | | Authors: | Wilke, M.S, Hills, T.L, Zhang, H.Z, Chambers, H.F, Strynadka, N.C. | | Deposit date: | 2004-08-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus.
J.Biol.Chem., 279, 2004
|
|
6Q9N
 
 | | Crystal structure of PBP2a from MRSA in complex with piperacillin and quinazolinone | | Descriptor: | 3-[2-[(~{E})-2-(4-ethynylphenyl)ethenyl]-4-oxidanylidene-quinazolin-3-yl]benzoic acid, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Martinez-Caballero, S, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Quinazolinone Allosteric Inhibitor of PBP 2a Synergizes with Piperacillin and Tazobactam against Methicillin-Resistant Staphylococcus aureus.
Antimicrob.Agents Chemother., 63, 2019
|
|
8AIK
 
 | |
8AKH
 
 | | Crystal structure of DltE from L. plantarum soaked with LTA | | Descriptor: | Beta-lactamase family protein, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Ravaud, S, Nikolopoulos, N, Grangeasse, C. | | Deposit date: | 2022-07-29 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analysis of Lactiplantibacillus plantarum DltE& reveals D-alanylated lipoteichoic acids as direct cues supporting Drosophila juvenile growth.
Elife, 12, 2023
|
|
8AGR
 
 | | Crystal structure of DltE from L. plantarum, apo form | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase family protein, SULFATE ION | | Authors: | Ravaud, S, Nikolopoulos, N, Grangeasse, C. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-function analysis of Lactiplantibacillus plantarum DltE& reveals D-alanylated lipoteichoic acids as direct cues supporting Drosophila juvenile growth.
Elife, 12, 2023
|
|
