6C23
 
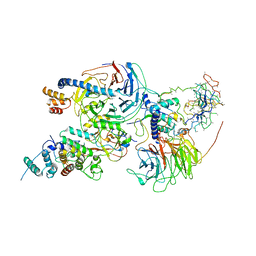 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Compact Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-05 | | Release date: | 2018-01-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
5IJ8
 
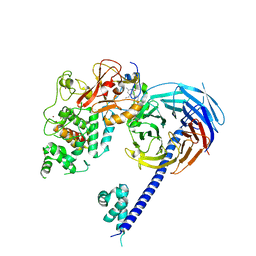 | | Structure of the primary oncogenic mutant Y641N Hs/AcPRC2 in complex with a pyridone inhibitor | | Descriptor: | 5,8-dichloro-2-[(4-ethyl-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-({1-[(2R)-2-hydroxypropanoyl]piperidin-4-yl}oxy)-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of Zeste Homolog 2 (EZH2),Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Deng, Y.-L, Liu, W. | | Deposit date: | 2016-03-01 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance.
Nat Commun, 7, 2016
|
|
6B3W
 
 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit,Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
6BX3
 
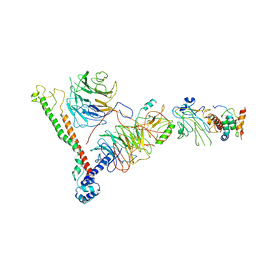 | | Structure of histone H3k4 methyltransferase | | Descriptor: | COMPASS component BRE2, COMPASS component SDC1, COMPASS component SPP1, ... | | Authors: | Skiniotis, G, Qu, Q.H. | | Deposit date: | 2017-12-16 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and Conformational Dynamics of a COMPASS Histone H3K4 Methyltransferase Complex.
Cell, 174, 2018
|
|
6C24
 
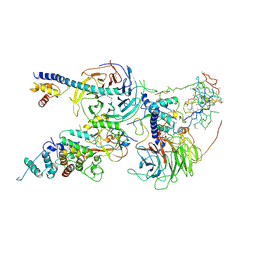 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-06 | | Release date: | 2018-01-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
6PWW
 
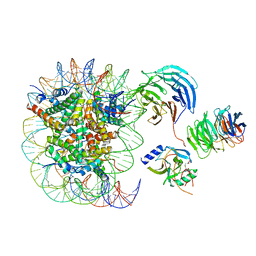 | | Cryo-EM structure of MLL1 in complex with RbBP5 and WDR5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
5WF7
 
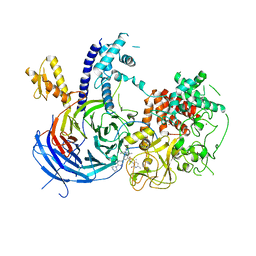 | | Chaetomium thermophilum Polycomb Repressive Complex 2 bound to GSK126 | | Descriptor: | 1-[(2S)-butan-2-yl]-N-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-indole-4-carboxamide, Histone-lysine-N-methyltransferase EZH2, Polycomb protein SUZ12 chimera, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-07-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Evolutionarily Conserved Structural Platform for PRC2 Inhibition by a Class of Ezh2 Inhibitors.
Sci Rep, 8, 2018
|
|
5WG6
 
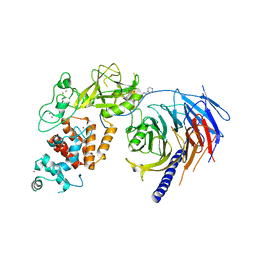 | | Human Polycomb Repressive Complex 2 in complex with GSK126 inhibitor | | Descriptor: | 1-[(2S)-butan-2-yl]-N-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-indole-4-carboxamide, Histone-lysine N-methyltransferase EZH2,Polycomb protein SUZ12 (E.C.2.1.1.43) chimera, Polycomb protein EED, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-07-13 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.901 Å) | | Cite: | An Evolutionarily Conserved Structural Platform for PRC2 Inhibition by a Class of Ezh2 Inhibitors.
Sci Rep, 8, 2018
|
|
5WFC
 
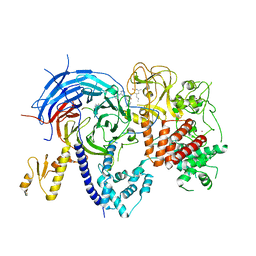 | |
5WFD
 
 | | Humanized mutant of the Chaetomium thermophilum Polycomb Repressive Complex 2 bound to the inhibitor GSK126 | | Descriptor: | 1-[(2S)-butan-2-yl]-N-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-indole-4-carboxamide, Histone-lysine-N-methyltransferase EZH2, Polycomb protein SUZ12 chimera, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-07-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | An Evolutionarily Conserved Structural Platform for PRC2 Inhibition by a Class of Ezh2 Inhibitors.
Sci Rep, 8, 2018
|
|
6CHG
 
 | | Crystal structure of the yeast COMPASS catalytic module | | Descriptor: | H3, Histone-lysine N-methyltransferase, H3 lysine-4 specific, ... | | Authors: | Hsu, P.L, Li, H, Zheng, N. | | Deposit date: | 2018-02-22 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.985 Å) | | Cite: | Crystal Structure of the COMPASS H3K4 Methyltransferase Catalytic Module.
Cell, 174, 2018
|
|
7W6L
 
 | | The crystal structure of MLL3-RBBP5-ASH2L in complex with H3K4me0 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6A
 
 | | Crystal structure of the MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L complex | | Descriptor: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6I
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me1 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6J
 
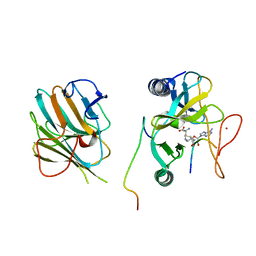 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me2 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W67
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me0 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7XPX
 
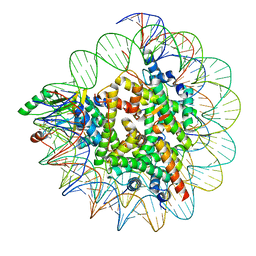 | |
7YRD
 
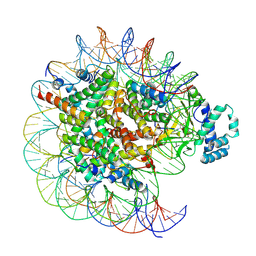 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
7YRG
 
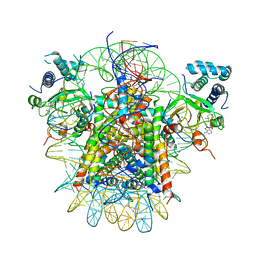 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
5TQR
 
 | |
