3HD0
 
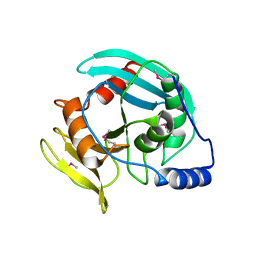 | | Crystal structure of Tm1865, an Endonuclease V from Thermotoga Maritima | | Descriptor: | Endonuclease V | | Authors: | Utepbergenov, D, Cooper, D.R, Derewenda, U, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-05-06 | | Release date: | 2009-07-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Tm1865, an Endonuclease V from Thermotoga Maritima
To be Published
|
|
6ES7
 
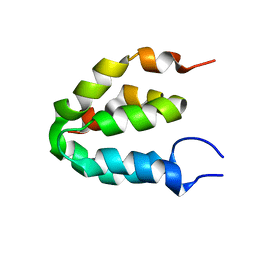 | |
7WUB
 
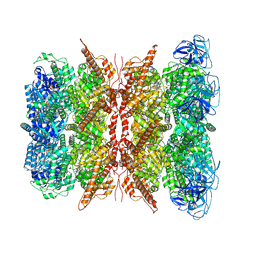 | | Cryo-EM structure of dodecamer P97 | | Descriptor: | 3-[3-cyclopentylsulfanyl-5-[[3-methyl-4-(4-methylsulfonylphenyl)phenoxy]methyl]-1,2,4-triazol-4-yl]pyridine, ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Liu, S, Wang, T. | | Deposit date: | 2022-02-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of dodecamer P97 at 2.99 Angstroms resolution
To Be Published
|
|
4FZV
 
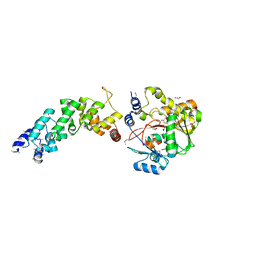 | | Crystal structure of the human MTERF4:NSUN4:SAM ternary complex | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Putative methyltransferase NSUN4, ... | | Authors: | Guja, K.E, Yakubovskaya, E, Mejia, E, Castano, S, Hambardjieva, E, Choi, W.S, Garcia-Diaz, M. | | Deposit date: | 2012-07-08 | | Release date: | 2012-10-03 | | Last modified: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (1.9996 Å) | | Cite: | Structure of the Essential MTERF4:NSUN4 Protein Complex Reveals How an MTERF Protein Collaborates to Facilitate rRNA Modification.
Structure, 20, 2012
|
|
6ES6
 
 | |
2EO0
 
 | |
3GA2
 
 | | Crystal structure of the Endonuclease_V (BSU36170) from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR624 | | Descriptor: | Endonuclease V | | Authors: | Forouhar, F, Abashidze, M, Hussain, M, Seetharaman, J, Janjua, H, Fang, Y, Xiao, R, Cunningham, K, Ma, L.-C, Owens, L, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-16 | | Release date: | 2009-02-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Endonuclease_V (BSU36170) from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR624
To be Published
|
|
4DQM
 
 | | Revealing a marine natural product as a novel agonist for retinoic acid receptors with a unique binding mode and antitumor activity | | Descriptor: | (5S)-4-[(3E,7E)-4,8-dimethyl-10-(2,6,6-trimethylcyclohex-1-en-1-yl)deca-3,7-dien-1-yl]-5-hydroxyfuran-2(5H)-one, Nuclear receptor coactivator 1, Retinoic acid receptor alpha | | Authors: | Wang, S, Wang, Z, Lin, S, Zheng, W, Wang, R, Jin, S, Chen, J, Jin, L, Li, Y. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Revealing a natural marine product as a novel agonist for retinoic acid receptors with a unique binding mode and inhibitory effects on cancer cells.
Biochem.J., 446, 2012
|
|
6ES5
 
 | |
6AOC
 
 | | Crystal Structure of an N-Hydroxythienopyrimidine-2,4-dione RNase H Active Site Inhibitor with Multiple Binding Modes to HIV Reverse Transcriptase | | Descriptor: | 1,2-ETHANEDIOL, 6-benzyl-3-hydroxythieno[2,3-d]pyrimidine-2,4(1H,3H)-dione, MANGANESE (II) ION, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-08-15 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, synthesis and biological evaluations of N-Hydroxy thienopyrimidine-2,4-diones as inhibitors of HIV reverse transcriptase-associated RNase H.
Eur J Med Chem, 141, 2017
|
|
2XK9
 
 | | Structural analysis of checkpoint kinase 2 (Chk2) in complex with inhibitor PV1533 | | Descriptor: | CHECKPOINT KINASE 2, N-{4-[(1E)-N-(N-hydroxycarbamimidoyl)ethanehydrazonoyl]phenyl}-7-nitro-1H-indole-2-carboxamide | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Shoemaker, R.H, Pommier, Y, Waugh, D.S. | | Deposit date: | 2010-07-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
6IAM
 
 | |
2JK3
 
 | |
5OGB
 
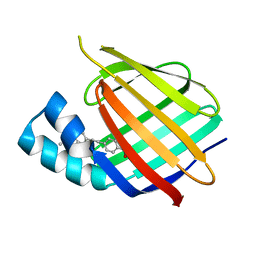 | | Human Cellular Retinoic Acid Binding Protein II (CRABPII) with bound synthetic retinoid DC360. | | Descriptor: | 4-[2-(4,4-dimethyl-1-propan-2-yl-quinolin-6-yl)ethynyl]benzoic acid, Cellular retinoic acid-binding protein 2 | | Authors: | Chisholm, D, Tomlinson, C, Whiting, A, Pohl, E. | | Deposit date: | 2017-07-12 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fluorescent Retinoic Acid Analogues as Probes for Biochemical and Intracellular Characterization of Retinoid Signaling Pathways.
Acs Chem.Biol., 14, 2019
|
|
3BYH
 
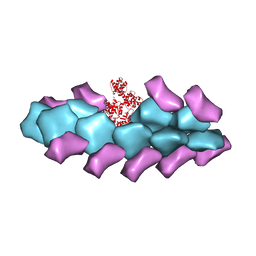 | | Model of actin-fimbrin ABD2 complex | | Descriptor: | Actin, fimbrin ABD2 | | Authors: | Galkin, V.E, Orlova, A, Cherepanova, O, Lebart, M.C, Egelman, E.H. | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | High-resolution cryo-EM structure of the F-actin-fimbrin/plastin ABD2 complex.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7X4U
 
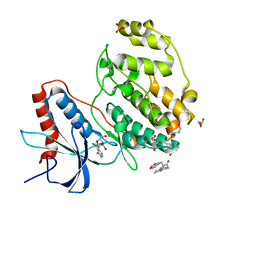 | | Crystal structure of ERK2 with an allosteric inhibitor 2 | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Yoshida, M, Kinoshita, T. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for producing allosteric ERK2 inhibitors
To Be Published
|
|
3D55
 
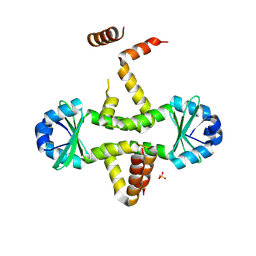 | | Crystal structure of M. tuberculosis YefM antitoxin | | Descriptor: | SULFATE ION, Uncharacterized protein Rv3357/MT3465 | | Authors: | Kumar, P, Issac, B, Dodson, E.J, Turkenberg, J.P, Mande, S.C. | | Deposit date: | 2008-05-15 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein
J.Mol.Biol., 383, 2008
|
|
7XC1
 
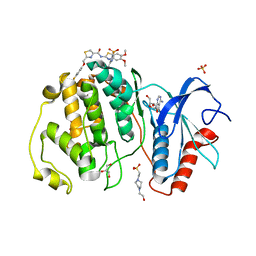 | | Crystal structure of ERK2 with an allosteric inhibitor 3 | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Yoshida, M, Kinoshita, T. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis for ERK2 allosteric inhibitors.
To Be Published
|
|
4DQ2
 
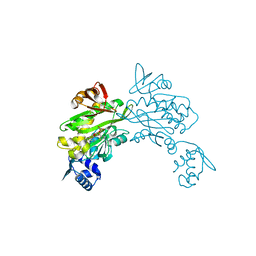 | | Structure of staphylococcus aureus biotin protein ligase in complex with biotinol-5'-amp | | Descriptor: | ((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHYL 5-((3AS,4S,6AR)-2-OXO-HEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL)PENTYL HYDROGEN PHOSPHATE, Biotin-[acetyl-CoA-carboxylase] ligase | | Authors: | Wilce, M, Yap, M, Pendini, N, Soares de Costa, T, Polyak, S, Tieu, W, Booker, G, Wallace, J. | | Deposit date: | 2012-02-14 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective inhibition of biotin protein ligase from Staphylococcus aureus.
J.Biol.Chem., 287, 2012
|
|
3CTO
 
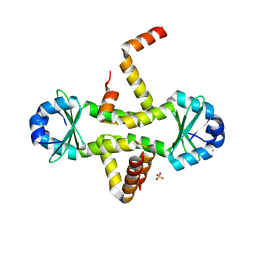 | | Crystal Structure of M. tuberculosis YefM antitoxin | | Descriptor: | SULFATE ION, Uncharacterized protein Rv3357/MT3465 | | Authors: | Kumar, P, Issac, B, Dodson, E.J, Turkenberg, J.P, Mande, S.C. | | Deposit date: | 2008-04-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein
J.Mol.Biol., 383, 2008
|
|
2Z1I
 
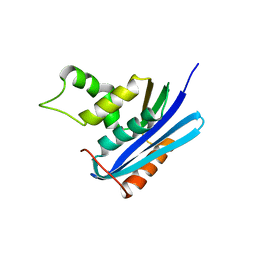 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K/Q113R/Q115K) | | Descriptor: | Ribonuclease HI | | Authors: | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
2Z1J
 
 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K/Q113R/Q115K/N143K/T145K) | | Descriptor: | Ribonuclease HI | | Authors: | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
5QXJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of ATAD2 in complex with PC578 | | Descriptor: | (3aS,8S,9aS)-10-methyl-4-oxo-1,4,6,8,9,9a-hexahydro-3a,8-epiminocyclohepta[1,2-c:4,5-c']dipyrrole-2(3H)-carbaldehyde, 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, ... | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | PanDDA analysis group deposition - Bromodomain of human ATAD2 fragment screening
To Be Published
|
|
5QXI
 
 | | PanDDA analysis group deposition -- Crystal Structure of ATAD2 in complex with PC587 | | Descriptor: | (4R,4aR,7aS,9R)-3,10-dimethyl-5,6,7,7a,8,9-hexahydro-4H-4a,9-epiminopyrrolo[3',4':5,6]cyclohepta[1,2-d][1,2]oxazol-4-ol, (4R,4aS,7aS,9S)-3,10-dimethyl-5,6,7,7a,8,9-hexahydro-4H-4a,9-epiminopyrrolo[3',4':5,6]cyclohepta[1,2-d][1,2]oxazol-4-ol, 1,2-ETHANEDIOL, ... | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | PanDDA analysis group deposition - Bromodomain of human ATAD2 fragment screening
To Be Published
|
|
5QXK
 
 | | PanDDA analysis group deposition -- Crystal Structure of ATAD2 in complex with PC581 | | Descriptor: | (3aS,8S,9aS)-2-acetyl-10-methyl-2,3,6,8,9,9a-hexahydro-3a,8-epiminocyclohepta[1,2-c:4,5-c']dipyrrol-4(1H)-one, 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, ... | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | PanDDA analysis group deposition - Bromodomain of human ATAD2 fragment screening
To Be Published
|
|
