1A16
 
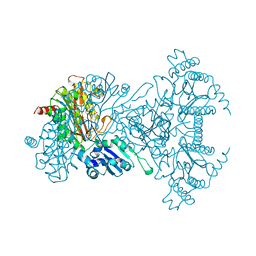 | | AMINOPEPTIDASE P FROM E. COLI WITH THE INHIBITOR PRO-LEU | | Descriptor: | AMINOPEPTIDASE P, LEUCINE, MANGANESE (II) ION, ... | | Authors: | Wilce, M.C, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1AV4
 
 | |
1AVK
 
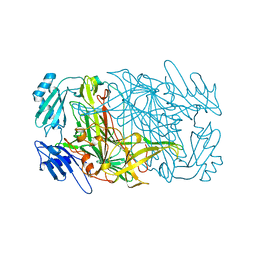
 | | CRYSTAL STRUCTURES OF THE COPPER-CONTAINING AMINE OXIDASE FROM ARTHROBACTER GLOBIFORMIS IN THE HOLO-AND APO-FORMS: IMPLICATIONS FOR THE BIOGENESIS OF TOPA QUINONE | | Descriptor: | AMINE OXIDASE | | Authors: | Wilce, M.C.J, Guss, J.M, Freeman, H.C, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 1997-09-16 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the copper-containing amine oxidase from Arthrobacter globiformis in the holo and apo forms: implications for the biogenesis of topaquinone.
Biochemistry, 36, 1997
|
|
1AVL
 
 | |
8ENI
 
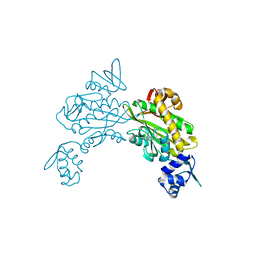 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | 3-[4-(5-fluoro-4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)butyl]-5-methyl-1,3-benzoxazol-2(3H)-one, Bifunctional ligase/repressor BirA | | Authors: | Wilce, M.C.J, Cini, D.A. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
4DQ2
 
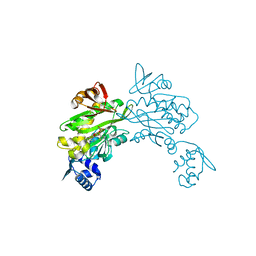 | | Structure of staphylococcus aureus biotin protein ligase in complex with biotinol-5'-amp | | Descriptor: | ((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHYL 5-((3AS,4S,6AR)-2-OXO-HEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL)PENTYL HYDROGEN PHOSPHATE, Biotin-[acetyl-CoA-carboxylase] ligase | | Authors: | Wilce, M, Yap, M, Pendini, N, Soares de Costa, T, Polyak, S, Tieu, W, Booker, G, Wallace, J. | | Deposit date: | 2012-02-14 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective inhibition of biotin protein ligase from Staphylococcus aureus.
J.Biol.Chem., 287, 2012
|
|
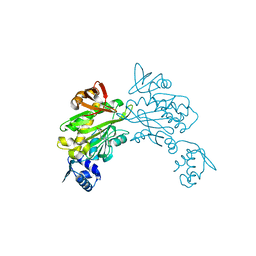
3RKY
 
 | |
1WVN
 
 | |
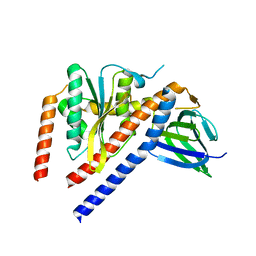
1FS0
 
 | |
3RKW
 
 | |
1JAW
 
 | | AMINOPEPTIDASE P FROM E. COLI LOW PH FORM | | Descriptor: | ACETATE ION, AMINOPEPTIDASE P, MANGANESE (II) ION | | Authors: | Wilce, M.C.J, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3RKX
 
 | |
1KSI
 
 | | CRYSTAL STRUCTURE OF A EUKARYOTIC (PEA SEEDLING) COPPER-CONTAINING AMINE OXIDASE AT 2.2A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Wilce, M.C.J, Kumar, V, Freeman, H.C, Guss, J.M. | | Deposit date: | 1996-07-20 | | Release date: | 1997-12-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a eukaryotic (pea seedling) copper-containing amine oxidase at 2.2 A resolution.
Structure, 4, 1996
|
|
3RIR
 
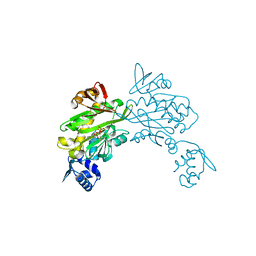 | |
6ORU
 
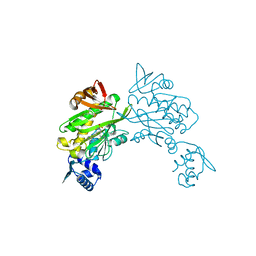 | |
7MP3
 
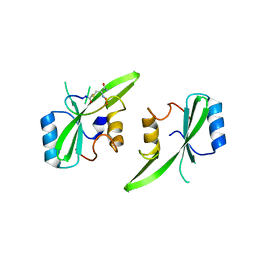 | |
5ITH
 
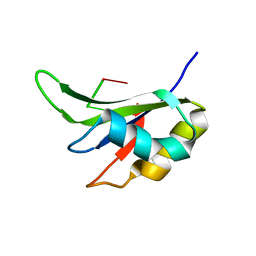 | |
5EEQ
 
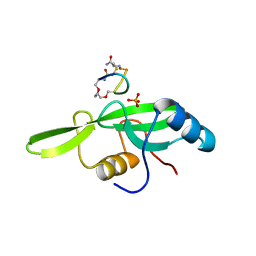 | | Grb7 SH2 with the G7-B1 bicyclic peptide inhibitor | | Descriptor: | Bicyclic Peptide Inhibitor, Growth factor receptor-bound protein 7, PHOSPHATE ION | | Authors: | Ambaye, N.D, Watson, G.M, Wilce, M.C.J, Wilce, G.M. | | Deposit date: | 2015-10-23 | | Release date: | 2016-06-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
4X6S
 
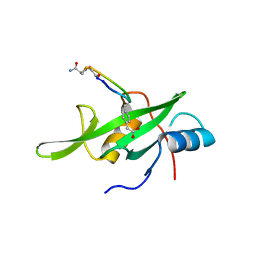 | | Grb7 SH2 domain with phosphotyrosine mimetic inhibitor peptide | | Descriptor: | Growth factor receptor-bound protein 7, Phosphotyrosine mimetic inhibitor peptide G7-TEM1 | | Authors: | Watson, G.M, Panjikar, S, Wilce, M.C, Wilce, J.A. | | Deposit date: | 2014-12-09 | | Release date: | 2015-09-23 | | Last modified: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Cyclic Peptides Incorporating Phosphotyrosine Mimetics as Potent and Specific Inhibitors of the Grb7 Breast Cancer Target.
J.Med.Chem., 58, 2015
|
|
1BE7
 
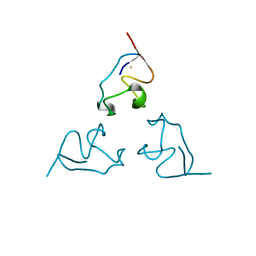 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN C42S MUTANT | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Maher, M, Guss, J.M, Wilce, M, Wedd, A.G. | | Deposit date: | 1998-05-20 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Rubredoxin from Clostridium Pasteurianum: Mutation of the Iron Cysteinyl Ligands to Serine. Crystal and Molecular Structures of the Oxidised and Dithionite-Treated Forms of the Cys42Ser Mutant
J.Am.Chem.Soc., 120, 1998
|
|
8SKK
 
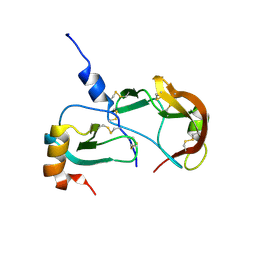 | | Crystal Structure of the Tick Evasin EVA-AAM1001(L39P) Complexed to Human Chemokine CCL17 | | Descriptor: | C-C motif chemokine 17, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Tick Evasin EVA-AAM1001(L39P) Complexed to Human Chemokine CCL7
To be Published
|
|
4WWQ
 
 | | Apo structure of the Grb7 SH2 domain | | Descriptor: | Growth factor receptor-bound protein 7, MALONIC ACID | | Authors: | Watson, G.M, Ambaye, N.D, Wilce, M.C, Wilce, J.A. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic Peptides Incorporating Phosphotyrosine Mimetics as Potent and Specific Inhibitors of the Grb7 Breast Cancer Target.
J.Med.Chem., 58, 2015
|
|
5EEL
 
 | | Grb7 SH2 with bicyclic peptide inhibitor | | Descriptor: | Bicyclic Peptide Inhibitor, FORMIC ACID, Growth factor receptor-bound protein 7, ... | | Authors: | Watson, G.M, Gunzburg, M.J, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2015-10-23 | | Release date: | 2016-06-15 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
5D0J
 
 | | Grb7 SH2 with inhibitor peptide | | Descriptor: | G7-TEdFP peptide, Growth factor receptor-bound protein 7, PHOSPHATE ION | | Authors: | Gunzburg, M.J, Watson, G.M, Wilce, J.A, Wilce, M.C.J. | | Deposit date: | 2015-08-03 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
1B13
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-26 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
