6G0J
 
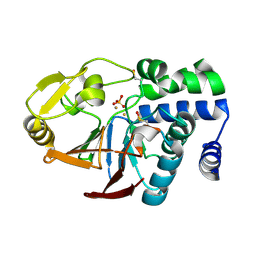 | | Inactive Fe-PP1 | | Descriptor: | FE (III) ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Salvi, F, Barabas, O, Koehn, M. | | Deposit date: | 2018-03-18 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effects of stably incorporated iron on protein phosphatase-1 structure and activity.
FEBS Lett., 592, 2018
|
|
5LMD
 
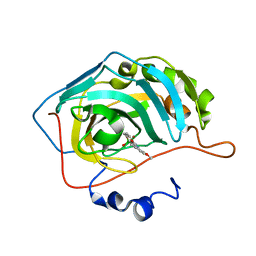 | | The crystal structure of hCA II in complex with a benzoxaborole inhibitor | | Descriptor: | 1-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-4-yl]-3-(2-methoxy-5-methyl-phenyl)urea, Carbonic anhydrase 2, ZINC ION | | Authors: | De Simone, G, Alterio, V, Esposito, D, Di Fiore, A. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Chem.Commun.(Camb.), 52, 2016
|
|
5LT6
 
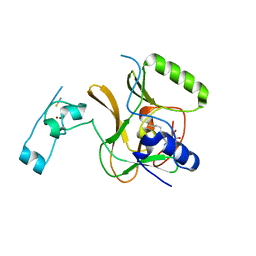 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, THIOCYANATE ION, ZINC ION, ... | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
6HI0
 
 | | Klebsiella pneumoniae Seryl-tRNA Synthetase in Complex with 5'-O-(N-(L-seryl)-Sulfamoyl)uridine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-seryl)-Sulfamoyl)uridine, CALCIUM ION, ... | | Authors: | Pang, L, De Graef, S, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2018-08-29 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
6G5C
 
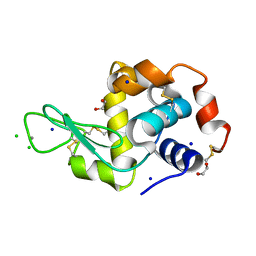 | | Interaction of TiO2 nanoparticles (NM-101) with Lysozyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Sbirkova-Dimitrova, H.I, Radoslavov, G, Hristov, P, Ganev, V, Dimowa, L, Rusev, R, Shivachev, B.L. | | Deposit date: | 2018-03-29 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallization and crystal structure of lysozyme in the presence of nanosized Titanium dioxide
Bul.Chem.Commn., 50, 2018
|
|
7ABI
 
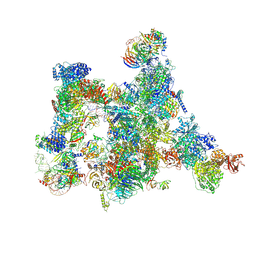 | | Human pre-Bact-2 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Beta-catenin-like protein 1, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2021-02-10 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
4Z4J
 
 | |
7Z74
 
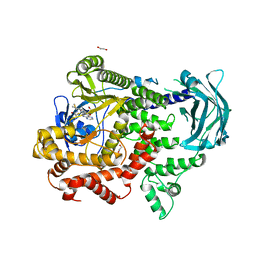 | | PI3KC2a core in complex with PITCOIN2 | | Descriptor: | 1,2-ETHANEDIOL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, ~{N}-[4-(3-hydroxyphenyl)-1,3-thiazol-2-yl]-2-[4-oxidanylidene-3-(2-phenylethyl)pteridin-2-yl]sulfanyl-ethanamide | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2022-03-15 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of selective inhibitors of phosphatidylinositol 3-kinase C2 alpha.
Nat.Chem.Biol., 19, 2023
|
|
7AGU
 
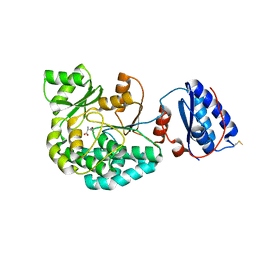 | | Structure of the S726F mutant of AcylTransferase domain of Mycocerosic Acid Synthase from Mycobacterium tuberculosis acylated with MethylMalonyl-coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, METHYLMALONIC ACID, Mycocerosic acid synthase | | Authors: | Brison, Y, Mourey, L, Maveyraud, L. | | Deposit date: | 2020-09-23 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Basis for Extender Unit Specificity of Mycobacterial Polyketide Synthases.
Acs Chem.Biol., 15, 2020
|
|
7Z75
 
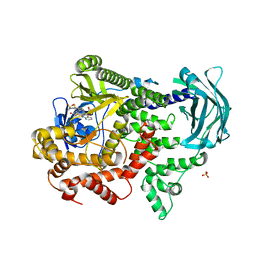 | | PI3KC2a core in complex with PITCOIN3 | | Descriptor: | 1,2-ETHANEDIOL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, SULFATE ION, ... | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2022-03-15 | | Release date: | 2022-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Development of selective inhibitors of phosphatidylinositol 3-kinase C2 alpha.
Nat.Chem.Biol., 19, 2023
|
|
6S52
 
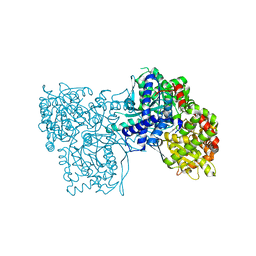 | | The crystal structure of glycogen phosphorylase in complex with 14 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-(5-phenyl-1,2,3,4-tetrazol-2-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-29 | | Release date: | 2020-02-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
3GVN
 
 | | The 1.2 Angstroem crystal structure of an E.coli tRNASer acceptor stem microhelix reveals two magnesium binding sites | | Descriptor: | 5'-R(*CP*CP*UP*CP*AP*CP*C)-3', 5'-R(*GP*GP*UP*GP*AP*GP*G)-3', MAGNESIUM ION | | Authors: | Eichert, A, Furste, J.P, Schreiber, A, Perbandt, M, Betzel, C, Erdmann, V.A, Forster, C. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2A crystal structure of an E. coli tRNASer)acceptor stem microhelix reveals two magnesium binding sites.
Biochem.Biophys.Res.Commun., 386, 2009
|
|
7AKG
 
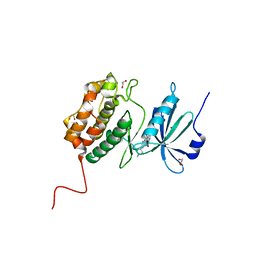 | |
7QSI
 
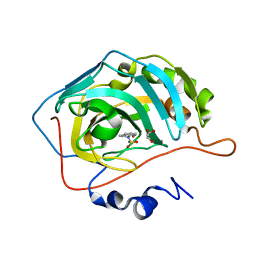 | | human Carbonic Anhydrase II in complex with indoline-1-sulfonamide | | Descriptor: | 2,3-dihydroindole-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-01-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Diversely substituted sulfamides for fragment-based drug discovery of carbonic anhydrase inhibitors: synthesis and inhibitory profile.
J Enzyme Inhib Med Chem, 37, 2022
|
|
8SSO
 
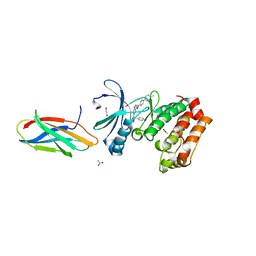 | | AurA bound to danusertib and inhibiting monobody Mb2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aurora kinase A, ... | | Authors: | Ludewig, H, Kim, C, Kern, D. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A biophysical framework for double-drugging kinases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BF3
 
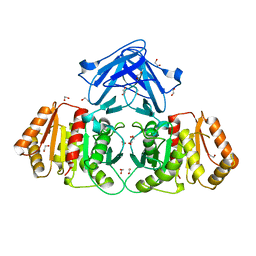 | |
5MIM
 
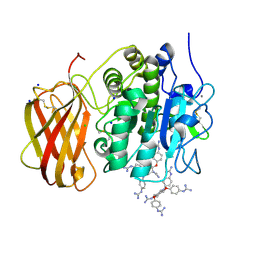 | | Xray structure of human furin bound with the 2,5-dideoxystreptamine derived small molecule inhibitor 1n | | Descriptor: | 1-[(1~{R},2~{R},4~{S},5~{S})-2,4-bis(4-carbamimidamidophenoxy)-5-[(4-carbamimidamidophenyl)amino]cyclohexyl]guanidine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Guan-Sheng, J, Than, M.E. | | Deposit date: | 2016-11-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies Revealed Active Site Distortions of Human Furin by a Small Molecule Inhibitor.
ACS Chem. Biol., 12, 2017
|
|
5YRX
 
 | | Crystal structure of a hypothetical protein Rv3716c from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Nucleoid-associated protein Rv3716c | | Authors: | Deka, G, Gopalan, A, Prabhavathi, M, Savithri, H.S, Raja, A, Murthy, M.R.N. | | Deposit date: | 2017-11-10 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biophysical characterization of Rv3716c, a hypothetical protein from Mycobacterium tuberculosis
Biochem. Biophys. Res. Commun., 495, 2018
|
|
6IHH
 
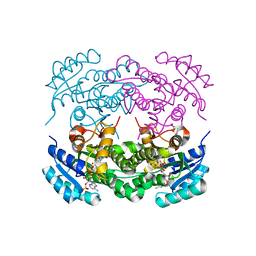 | | Crystal structure of RasADH F12 from Ralstonia.sp in complex with NADPH and A6O | | Descriptor: | (2R,3S)-2-ethyl-2-[(2E)-2-(6-methoxy-3,4-dihydro-2H-naphthalen-1-ylidene)ethyl]-3-oxidanyl-cyclopentan-1-one, Alclohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, H.L, Chen, X, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2018-09-30 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efficient reductive desymmetrization of bulky 1,3-cyclodiketones enabled by structure-guided directed evolution of a carbonyl reductase.
Nat Catal, 2, 2019
|
|
8PH5
 
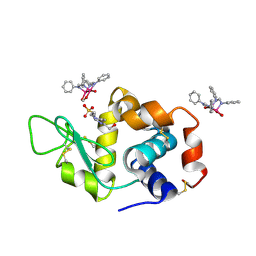 | | X-ray structure of the adduct formed upon reaction of Lysozyme with [Ru2Cl(DPhF)2(O2CCH3)2] in condition B | | Descriptor: | 1-oxidanyl-6,8,9,11-tetraphenyl-2,4-dioxa-6,8,9,11-tetraza-1$l^{5},5$l^{4}-diruthenatricyclo[3.3.3.0^{1,5}]undecane, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lysozyme C | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-19 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Steric hindrance and charge influence on the cytotoxic activity and protein binding properties of diruthenium complexes.
Int.J.Biol.Macromol., 253, 2023
|
|
8PX2
 
 | |
6VCW
 
 | | Crystal structure of Medicago truncatula S-adenosylmethionine Synthase 3A (MtMAT3A) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
6XYY
 
 | | Update of ACHE FROM DROSOPHILA MELANOGASTER COMPLEX WITH TACRINE DERIVATIVE 9-(3-PHENYLMETHYLAMINO)-1,2,3,4-TETRAHYDROACRIDINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 9-(3-PHENYLMETHYLAMINO)-1,2,3,4-TETRAHYDROACRIDINE, Acetylcholinesterase, ... | | Authors: | Nachon, F, Sussman, J.L. | | Deposit date: | 2020-01-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Second Look at the Crystal Structures ofDrosophila melanogasterAcetylcholinesterase in Complex with Tacrine Derivatives Provides Insights Concerning Catalytic Intermediates and the Design of Specific Insecticides.
Molecules, 25, 2020
|
|
7CAJ
 
 | | Crystal structure of SETDB1 Tudor domain in complexed with Compound 2. | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-5H-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Guo, Y.P, Liang, X, Xin, M, Luyi, H, Chengyong, W, Yang, S.Y. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
9OA8
 
 | |
