8RUQ
 
 | |
8F30
 
 | | LSD1-CoREST in complex with AW2, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-{5-[3-([1,1'-biphenyl]-4-yl)propanoyl]-7,8-dimethyl-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl}-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-11-09 | | Release date: | 2024-06-12 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Covalent adduct Grob fragmentation underlies LSD1 demethylase-specific inhibitor mechanism of action and resistance.
Nat Commun, 16, 2025
|
|
8F59
 
 | | LSD1-CoREST in complex with AW2 and SNAG peptide | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, Zinc finger protein SNAI1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-11-12 | | Release date: | 2024-06-12 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Covalent adduct Grob fragmentation underlies LSD1 demethylase-specific inhibitor mechanism of action and resistance.
Nat Commun, 16, 2025
|
|
8FJ4
 
 | | LSD1-CoREST in complex with T108, short soaking | | Descriptor: | 3-[(1R,2S)-2-(cyclobutylamino)cyclopropyl]-N-phenylbenzamide, Lysine-specific histone demethylase 1A, REST corepressor 1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-12-19 | | Release date: | 2024-06-26 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Covalent adduct Grob fragmentation underlies LSD1 demethylase-specific inhibitor mechanism of action and resistance.
Nat Commun, 16, 2025
|
|
8RUP
 
 | |
8F6S
 
 | | LSD1-CoREST in complex with T105 | | Descriptor: | 3-[(1R,2S)-2-(cyclobutylamino)cyclopropyl]-N-(5-methyl-1,3,4-thiadiazol-2-yl)benzamide, Lysine-specific histone demethylase 1A, REST corepressor 1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Covalent adduct Grob fragmentation underlies LSD1 demethylase-specific inhibitor mechanism of action and resistance.
Nat Commun, 16, 2025
|
|
5KA1
 
 | | Protein Tyrosine Phosphatase 1B Delta helix 7 mutant in complex with TCS401, closed state | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
8FJ7
 
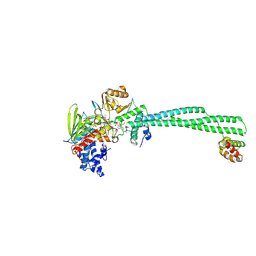 | | LSD1-CoREST in complex with T108 and SNAG peptide | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, Zinc finger protein SNAI1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-12-19 | | Release date: | 2024-06-26 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Covalent adduct Grob fragmentation underlies LSD1 demethylase-specific inhibitor mechanism of action and resistance.
Nat Commun, 16, 2025
|
|
6UCS
 
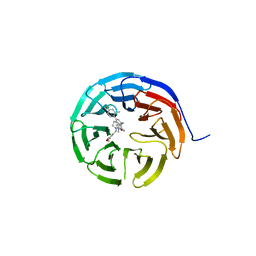 | |
6UDY
 
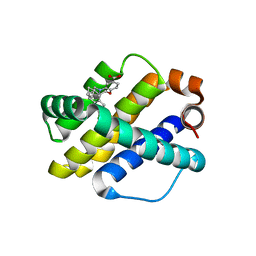 | | X-ray co-crystal structure of compound 5 with Mcl-1 | | Descriptor: | (3S)-6'-chloro-5-(cyclobutylmethyl)-3',4,4',5-tetrahydro-2H,2'H-spiro[1,5-benzoxazepine-3,1'-naphthalene]-7-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-09-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and in Vivo Evaluation of Macrocyclic Mcl-1 Inhibitors Featuring an alpha-Hydroxy Phenylacetic Acid Pharmacophore or Bioisostere.
J.Med.Chem., 62, 2019
|
|
6MGL
 
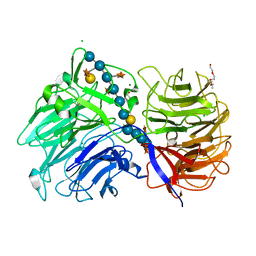 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, D60A mutant in complex with XXLG and XGXXLG xyloglucan | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Arnal, G, Watanabe, N, Brumer, H, Savchenko, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
8IOK
 
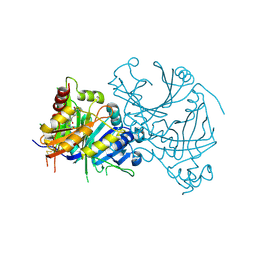 | | Crystal structure of AtHPPD-CLJ507 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 6-[diethylcarbamoyl(methyl)amino]-N-(1-methyl-1,2,3,4-tetrazol-5-yl)-2-(trifluoromethyl)pyridine-3-carboxamide, COBALT (II) ION | | Authors: | Dong, J, Yang, G.-F, Lin, H.-Y. | | Deposit date: | 2023-03-11 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Crystal structure of human HPPD-NTBC complex
To Be Published
|
|
6MF9
 
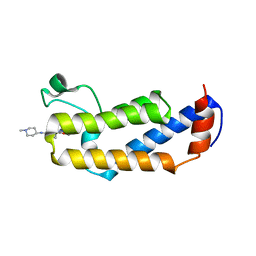 | | Crystal structure of CGD4-650 with compound BI2536 | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, ZnKn (C2HC)+Athook+bromo domain protein, Taf250, ... | | Authors: | Dong, A, Lin, Y.L, Hou, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-10 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.037 Å) | | Cite: | Crystal structure of CGD4-650 with compound BI2536
to be published
|
|
5XZR
 
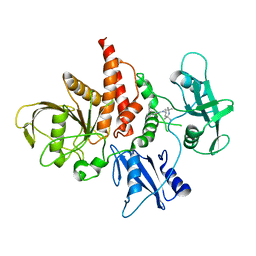 | | The atomic structure of SHP2 E76A mutant in complex with allosteric inhibitor 9b | | Descriptor: | 4-(3-phenylphenyl)-N-(2,2,6,6-tetramethylpiperidin-4-yl)-1,3-thiazol-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Li, D, Xie, J, Zhu, J, Liu, C. | | Deposit date: | 2017-07-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric Inhibitors of SHP2 with Therapeutic Potential for Cancer Treatment.
J. Med. Chem., 60, 2017
|
|
3ZWV
 
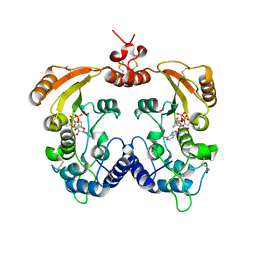 | | Crystal structure of ADP-ribosyl cyclase complexed with ara-2'F-ADP- ribose at 2.3 angstrom | | Descriptor: | ADP-RIBOSYL CYCLASE, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of intermediates along the cyclization pathway of Aplysia ADP-ribosyl cyclase.
J. Mol. Biol., 415, 2012
|
|
5KAG
 
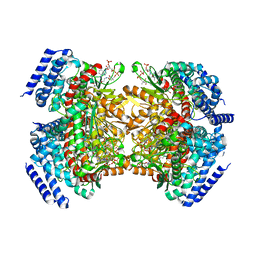 | | Crystal structure of a dioxygenase in the Crotonase superfamily in P21 | | Descriptor: | (3,5-dihydroxyphenyl)acetyl-CoA 1,2-dioxygenase, OXYGEN MOLECULE, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Li, K, Fielding, E.N, Condurso, H.L, Bruner, S.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | Probing the structural basis of oxygen binding in a cofactor-independent dioxygenase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6RD2
 
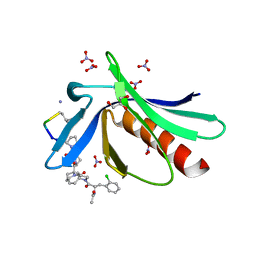 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-1]-TEDEL-NH2 | | Descriptor: | (3~{S},7~{R},10~{R},13~{S})-4-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, GLYCEROL, NITRATE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2019-04-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5Y4H
 
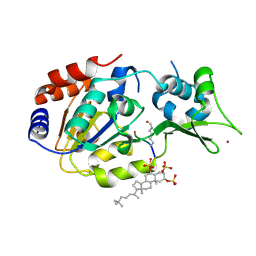 | | Human SIRT3 in complex with halistanol sulfate | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, THr-Arg-Ser-GLY-ALY-VAL-MET-ARG-ARG-LEU-ARG-ARG, ... | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2017-08-03 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Halistanol sulfates I and J, new SIRT1-3 inhibitory steroid sulfates from a marine sponge of the genus Halichondria
J. Antibiot., 71, 2018
|
|
6P38
 
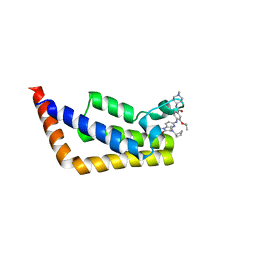 | | Crystal Structure Analysis of TAF1 Bromodomain | | Descriptor: | 4-{[(3R)-4-cyclopentyl-1,3-dimethyl-2-oxo-1,2,3,4-tetrahydropyrido[2,3-b]pyrazin-6-yl]amino}-N-(1-methylpiperidin-4-yl)-3-[(propan-2-yl)oxy]benzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual Inhibition of TAF1 and BET Bromodomains from the BI-2536 Kinase Inhibitor Scaffold.
Acs Med.Chem.Lett., 10, 2019
|
|
8X4N
 
 | | Crystal structure of the PI3P-binding domain of Legionella SetA in complex with inositol 1,3-bisphosphate | | Descriptor: | GLYCEROL, PHOSPHORIC ACID MONO-(2,3,4,6-TETRAHYDROXY-5-PHOSPHONOOXY-CYCLOHEXYL) ESTER, Subversion of eukaryotic traffic protein A | | Authors: | Im, H.N, Lee, Y, Jang, D.M, Hahn, H, Kim, H.S. | | Deposit date: | 2023-11-15 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of the PI3P-binding domain of Legionella SetA in complex with inositol 1,3-bisphosphate
To Be Published
|
|
5H2R
 
 | | Crystal structure of T brucei phosphodiesterase B2 bound to compound 15b | | Descriptor: | (4~{a}~{S},8~{a}~{R})-2-cycloheptyl-4-[4-(2-hydroxyethyloxy)-3-[2-(2-oxidanylideneimidazolidin-1-yl)ethoxy]phenyl]-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, MAGNESIUM ION, Phosphodiesterase, ... | | Authors: | Noble, C.G. | | Deposit date: | 2016-10-17 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trypanosomal Phosphodiesterase B1 and B2 as a Potential Therapy for Human African Trypanosomiasis
To Be Published
|
|
8X6L
 
 | | PRMT5:MEP50 WITH SCR-6920 AND SAM | | Descriptor: | 1-ethyl-8-[(2-methoxy-7-azaspiro[3.5]nonan-7-yl)carbonyl]-4-[2-oxidanylidene-2-[(3~{S})-1,2,3,4-tetrahydroisoquinolin-3-yl]ethyl]-2,3-dihydro-1,4-benzodiazepin-5-one, GLYCEROL, Methylosome protein WDR77, ... | | Authors: | Zhou, F, Zhou, F. | | Deposit date: | 2023-11-21 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | PRMT5:MEP50 WITH SCR-6920 AND SAM
To Be Published
|
|
6M7X
 
 | | Structure of human CYP11B1 in complex with fadrozole | | Descriptor: | 4-[(5S)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-5-yl]benzonitrile, Cytochrome P450 11B1, mitochondrial, ... | | Authors: | Scott, E.E, Brixius-Anderko, S. | | Deposit date: | 2018-08-21 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Structure of human cortisol-producing cytochrome P450 11B1 bound to the breast cancer drug fadrozole provides insights for drug design.
J. Biol. Chem., 294, 2019
|
|
8X7G
 
 | | Crystal structure of the ternary complex of GID4-PROTAC(NEP108)-BRD4(BD1). | | Descriptor: | 2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]-~{N}-[2-[2-[[2-[4-[2-(1~{H}-indol-2-ylmethylamino)ethanoylamino]cyclohexyl]-3~{H}-benzimidazol-5-yl]oxy]ethoxy]ethyl]ethanamide, Bromodomain-containing protein 4, Glucose-induced degradation protein 4 homolog | | Authors: | Dong, C, Yan, X, Li, Y. | | Deposit date: | 2023-11-24 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 108
To Be Published
|
|
6MGJ
 
 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Nocek, B, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
