3R34
 
 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant E73D complexed with CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxybenzoyl-CoA thioesterase, COENZYME A | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
3R3F
 
 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant T77A complexed with 4-hydroxyphenacyl CoA | | Descriptor: | 4-HYDROXYPHENACYL COENZYME A, 4-hydroxybenzoyl-CoA thioesterase | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
3R44
 
 | | Mycobacterium tuberculosis fatty acyl CoA synthetase | | Descriptor: | HISTIDINE, MALONATE ION, fatty acyl CoA synthetase FADD13 (FATTY-ACYL-CoA SYNTHETASE) | | Authors: | Andersson, C.S, Martinez Molina, D, Hogbom, M. | | Deposit date: | 2011-03-17 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Mycobacterium tuberculosis Very-Long-Chain Fatty Acyl-CoA Synthetase: Structural Basis for Housing Lipid Substrates Longer than the Enzyme.
Structure, 20, 2012
|
|
3R4S
 
 | | Cell entry of botulinum neurotoxin type C is dependent upon interaction with two ganglioside molecules | | Descriptor: | Botulinum neurotoxin type C1, N-acetyl-alpha-neuraminic acid, N-acetyl-beta-neuraminic acid | | Authors: | Strotmeier, J, Gu, S, Jutzi, S, Mahrhold, S, Zhou, J, Pich, A, Bigalke, H, Rummel, A, Jin, R, Binz, T. | | Deposit date: | 2011-03-17 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The biological activity of botulinum neurotoxin type C is dependent upon novel types of ganglioside binding sites.
Mol.Microbiol., 81, 2011
|
|
6UST
 
 | | Gut microbial sulfatase from Hungatella hathewayi | | Descriptor: | CALCIUM ION, N-acetylgalactosamine 6-sulfate sulfatase | | Authors: | Ervin, S.M, Redinbo, M.R. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into Endobiotic Reactivation by Human Gut Microbiome-Encoded Sulfatases.
Biochemistry, 59, 2020
|
|
7W2Z
 
 | | Cryo-EM structure of the ghrelin-bound human ghrelin receptor-Go complex | | Descriptor: | Appetite-regulating hormone, CHOLESTEROL, Growth hormone secretagogue receptor type 1, ... | | Authors: | Qin, J, Ming, Q, Ji, S, Mao, C, Shen, D, Zhang, Y. | | Deposit date: | 2021-11-24 | | Release date: | 2022-01-19 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular mechanism of agonism and inverse agonism in ghrelin receptor.
Nat Commun, 13, 2022
|
|
3R4U
 
 | | Cell entry of botulinum neurotoxin type C is dependent upon interaction with two ganglioside molecules | | Descriptor: | Botulinum neurotoxin type C1 | | Authors: | Strotmeier, J, Gu, S, Jutzi, S, Mahrhold, S, Zhou, J, Pich, A, Bigalke, H, Rummel, A, Jin, R, Binz, T. | | Deposit date: | 2011-03-17 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The biological activity of botulinum neurotoxin type C is dependent upon novel types of ganglioside binding sites.
Mol.Microbiol., 81, 2011
|
|
6MOA
 
 | | C-terminal bromodomain of human BRD2 in complex with 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole inhibitor | | Descriptor: | 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole, Bromodomain-containing protein 2, GLYCEROL | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MSM
 
 | | Phosphorylated, ATP-bound human cystic fibrosis transmembrane conductance regulator (CFTR) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, ... | | Authors: | Zhang, Z, Liu, F, Chen, J. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular structure of the ATP-bound, phosphorylated human CFTR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6MO7
 
 | | N-terminal bromodomain of human BRD2 with N-((4-(3-(N-cyclopentylsulfamoyl)-4-methylphenyl)-3-methylisoxazol-5-yl)methyl)acetamide inhibitor | | Descriptor: | Bromodomain-containing protein 2, N-({4-[3-(cyclopentylsulfamoyl)-4-methylphenyl]-3-methyl-1,2-oxazol-5-yl}methyl)acetamide | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
3QYE
 
 | | Crystal Structure of Human TBC1D1 RabGAP domain | | Descriptor: | TBC1 domain family member 1 | | Authors: | Park, S.Y, Shoelson, S.E. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human TBC1D1 and TBC1D4 (AS160) RabGTPase-activating protein (RabGAP) domains reveal critical elements for GLUT4 translocation.
J.Biol.Chem., 286, 2011
|
|
4C9Z
 
 | | Crystal structure of Siah1 at 1.95 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, E3 UBIQUITIN-PROTEIN LIGASE SIAH1, ... | | Authors: | Rimsa, V, Eadsforth, T.C, Hunter, W.N. | | Deposit date: | 2013-10-04 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Two High-Resolution Structures of the Human E3 Ubiquitin Ligase Siah1.
Acta Crystallogr.,Sect.F, 96, 2013
|
|
4CBN
 
 | |
3R00
 
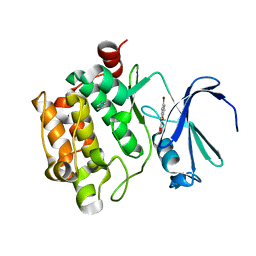 | |
3R35
 
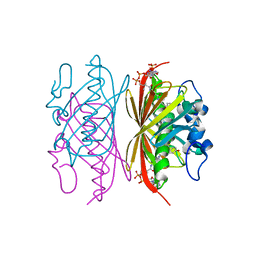 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant E73D complexed with 4-hydroxyphenacyl CoA | | Descriptor: | 4-HYDROXYPHENACYL COENZYME A, 4-hydroxybenzoyl-CoA thioesterase | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
8DHA
 
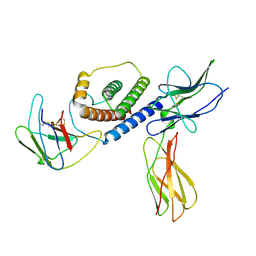 | |
8DH8
 
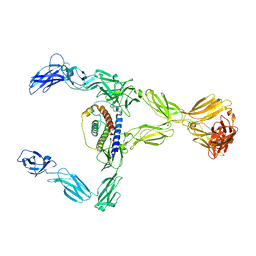 | |
6V1C
 
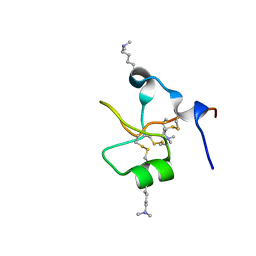 | | Crystal structure of human trefoil factor 3 in complex with its cognate ligand | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, Trefoil factor 3 | | Authors: | Jarva, M.A, Lingford, J.P, John, A, Scott, N.E, Goddard-Borger, E.D. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Trefoil factors share a lectin activity that defines their role in mucus.
Nat Commun, 11, 2020
|
|
4LHY
 
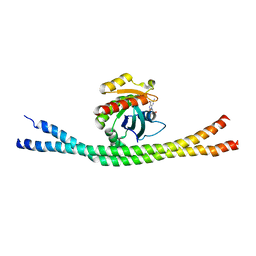 | | Crystal structure of GDP-bound Rab8:Rabin8 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Rab-3A-interacting protein, Ras-related protein Rab-8A, ... | | Authors: | Guo, Z, Hou, X.M, Goody, R.S, Itzen, A. | | Deposit date: | 2013-07-01 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Intermediates in the Guanine Nucleotide Exchange Reaction of Rab8 Protein Catalyzed by Guanine Nucleotide Exchange Factors Rabin8 and GRAB.
J.Biol.Chem., 288, 2013
|
|
7QE7
 
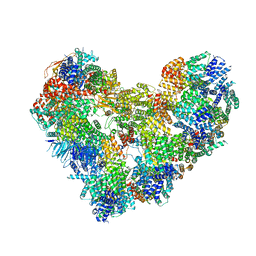 | | High-resolution structure of the Anaphase-promoting complex/cyclosome (APC/C) bound to co-activator Cdh1 | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Hoefler, A, Yu, J, Chang, L, Zhang, Z, Yang, J, Boland, A, Barford, D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-01-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution structure of the Anaphase-promoting complex (APC/C) bound to co-activator Cdh1
To Be Published
|
|
4LHZ
 
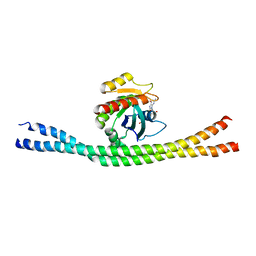 | | Crystal structure of GTP-bound Rab8:Rabin8 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Rab-3A-interacting protein, Ras-related protein Rab-8A | | Authors: | Guo, Z, Hou, X.M, Goody, R.S, Itzen, A. | | Deposit date: | 2013-07-01 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Intermediates in the Guanine Nucleotide Exchange Reaction of Rab8 Protein Catalyzed by Guanine Nucleotide Exchange Factors Rabin8 and GRAB.
J.Biol.Chem., 288, 2013
|
|
3TCU
 
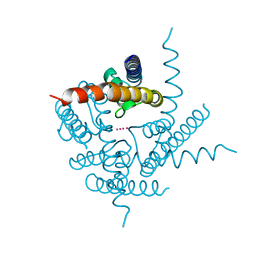 | | Crystal Structure of NaK2K Channel D68E Mutant | | Descriptor: | POTASSIUM ION, Potassium channel protein | | Authors: | Sauer, D.B, Zeng, W, Raghunathan, S, Jiang, Y. | | Deposit date: | 2011-08-09 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6N55
 
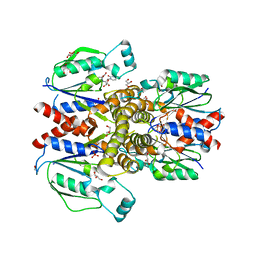 | | Crystal structure of human uridine-cytidine kinase 2 complexed with 2'-azidouridine | | Descriptor: | 2'-azido-2'-deoxyuridine, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Cuthbert, B.J, Nainar, S, Spitale, R.C, Goulding, C.W. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.085 Å) | | Cite: | An optimized chemical-genetic method for cell-specific metabolic labeling of RNA.
Nat.Methods, 17, 2020
|
|
7WMV
 
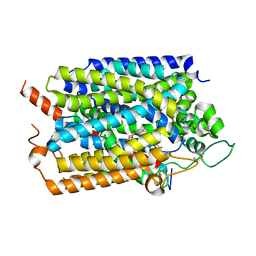 | | Structure of human SGLT1-MAP17 complex bound with LX2761 | | Descriptor: | N-[2-(dimethylamino)ethyl]-2-methyl-2-[4-[4-[[2-methyl-5-[(2S,3R,4R,5S,6R)-6-methylsulfanyl-3,4,5-tris(oxidanyl)oxan-2-yl]phenyl]methyl]phenyl]butanoylamino]propanamide, PDZK1-interacting protein 1, Sodium/glucose cotransporter 1 | | Authors: | Chen, L, Niu, Y, Cui, W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of SGLT1 inhibitors.
Nat Commun, 13, 2022
|
|
3R02
 
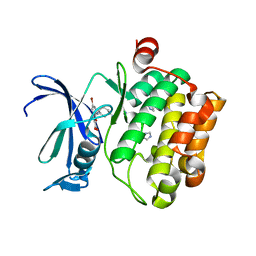 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 7-[(cis-4-aminocyclohexyl)amino]-5-bromo-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
