3JSV
 
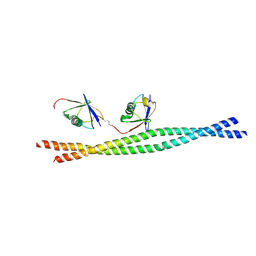 | | Crystal structure of mouse NEMO CoZi in complex with Lys63-linked di-ubiquitin | | Descriptor: | NF-kappa-B essential modulator, Ubiquitin | | Authors: | Yoshikawa, A, Sato, Y, Mimura, H, Yamashita, M, Yamagata, A, Fukai, S. | | Deposit date: | 2009-09-11 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63-linked di-ubiquitin
Febs Lett., 583, 2009
|
|
7M3Q
 
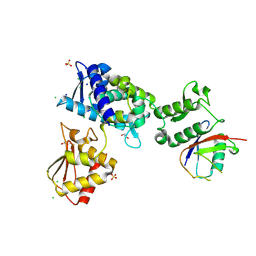 | | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, ... | | Authors: | Chowdhury, A, Singer, A.U, Ogunjimi, A.A, Teyra, J, Zhang, W, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-03-18 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV)
To be published
|
|
5N2W
 
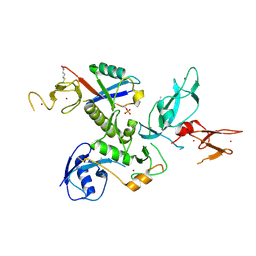 | | WT-Parkin and pUB complex | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, Polyubiquitin-B, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5LRX
 
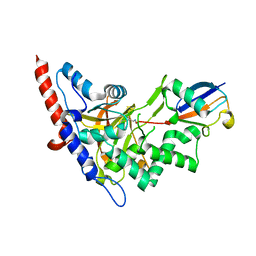 | | Structure of A20 OTU domain bound to ubiquitin | | Descriptor: | Polyubiquitin-B, Tumor necrosis factor alpha-induced protein 3 | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
3I3T
 
 | | Crystal structure of covalent ubiquitin-USP21 complex | | Descriptor: | ETHANAMINE, Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 21, ... | | Authors: | Neculai, D, Avvakumov, G.V, Walker, J.R, Xue, S, Butler-Cole, C, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
5N38
 
 | | S65DParkin and pUB complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5LRV
 
 | | Structure of Cezanne/OTUD7B OTU domain bound to Lys11-linked diubiquitin | | Descriptor: | GLYCEROL, OTU domain-containing protein 7B, PHOSPHATE ION, ... | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
3KVF
 
 | |
3KW5
 
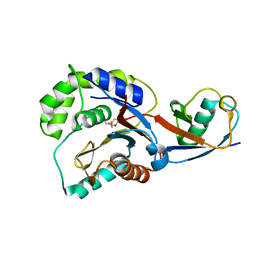 | |
7O2W
 
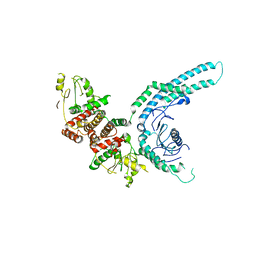 | | Structure of the C9orf72-SMCR8 complex | | Descriptor: | Guanine nucleotide exchange protein SMCR8,Guanine nucleotide exchange protein SMCR8,Maltose/maltodextrin-binding periplasmic protein, Ubiquitin-like protein SMT3,Guanine nucleotide exchange C9orf72 | | Authors: | Noerpel, J, Cavadini, S, Schenk, A.D, Graff-Meyer, A, Chao, J, Bhaskar, V. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structure of the human C9orf72-SMCR8 complex reveals a multivalent protein interaction architecture.
Plos Biol., 19, 2021
|
|
3L0W
 
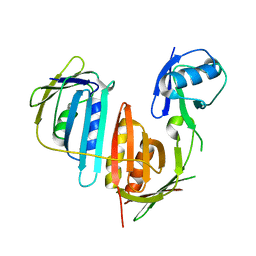 | | Structure of split monoubiquitinated PCNA with ubiquitin in position two | | Descriptor: | Monoubiquitinated Proliferating cell nuclear antigen, Proliferating cell nuclear antigen | | Authors: | Freudenthal, B.D, Gakhar, L, Ramaswamy, S, Washington, M.T. | | Deposit date: | 2009-12-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of monoubiquitinated PCNA and implications for translesion synthesis and DNA polymerase exchange.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7OWD
 
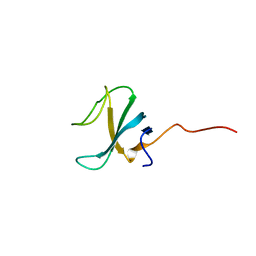 | |
7OWC
 
 | |
7OJE
 
 | | Crystal structure of the covalent complex between Tribolium castaneum deubiquitinase ZUP and Ubiquitin-PA | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Lys-63-specific deubiquitinase ZUFSP, ... | | Authors: | Pichlo, C, Hermanns, T, Hofmann, K, Baumann, U. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structural basis for the diverse linkage specificities within the ZUFSP deubiquitinase family.
Nat Commun, 13, 2022
|
|
6EI1
 
 | | Crystal structure of the covalent complex between deubiquitinase ZUFSP (ZUP1) and Ubiquitin-PA | | Descriptor: | GLYCEROL, MALONATE ION, Polyubiquitin-B, ... | | Authors: | Pichlo, C, Baumann, U, Hofmann, K, Hermanns, T. | | Deposit date: | 2017-09-16 | | Release date: | 2018-03-07 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | A family of unconventional deubiquitinases with modular chain specificity determinants.
Nat Commun, 9, 2018
|
|
7P7I
 
 | | Native structure of N-acetylglucosamine kinase from Plesiomonas shigelloides | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P7W
 
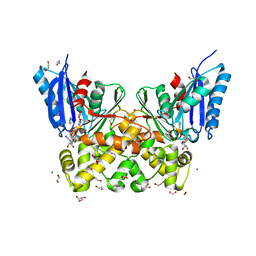 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7PI4
 
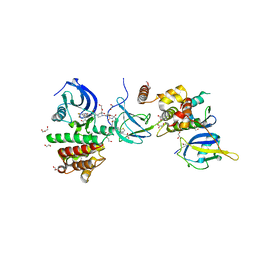 | | FAK Protac GSK215 in complex with FAK and pVHL:ElonginC:ElonginB | | Descriptor: | (2S,4R)-4-hydroxy-1-((S)-2-(2-(4-(3-methoxy-4-((4-((2-(methylcarbamoyl)phenyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)phenyl)piperazin-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Chung, C. | | Deposit date: | 2021-08-19 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery and Characterisation of Highly Cooperative FAK-Degrading PROTACs.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7PLO
 
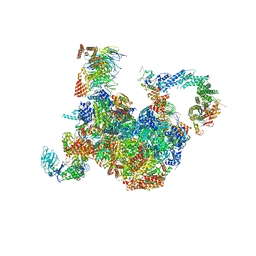 | | H. sapiens replisome-CUL2/LRR1 complex | | Descriptor: | Cell division control protein 45 homolog, Claspin, Cullin-2, ... | | Authors: | Jones, M.J, Yeeles, J.T.P, Deegan, T.D, Jenkyn-Bedford, M. | | Deposit date: | 2021-09-01 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A conserved mechanism for regulating replisome disassembly in eukaryotes.
Nature, 600, 2021
|
|
7PYV
 
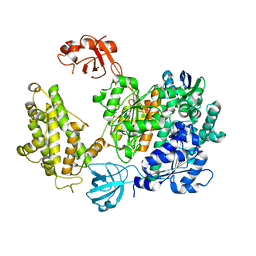 | | Crystal structure of human UBA6 in complex with the ubiquitin-like modifier FAT10 | | Descriptor: | UBD, Ubiquitin-like modifier-activating enzyme 6,Ubiquitin-like modifier-activating enzyme 1,Ubiquitin-like modifier-activating enzyme 6 | | Authors: | Li, S, Truongvan, N, Schindelin, H. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
6FGE
 
 | | Crystal structure of human ZUFSP/ZUP1 in complex with ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, AMMONIUM ION, ... | | Authors: | Kwasna, D, Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2018-01-10 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery and Characterization of ZUFSP/ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability.
Mol. Cell, 70, 2018
|
|
6FDK
 
 | |
6FFA
 
 | | FMDV Leader protease bound to substrate ISG15 | | Descriptor: | GLYCEROL, Lbpro, SULFATE ION, ... | | Authors: | Swatek, K.N, Pruneda, J.N, Komander, D. | | Deposit date: | 2018-01-05 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Irreversible inactivation of ISG15 by a viral leader protease enables alternative infection detection strategies.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FYH
 
 | | Disulfide between ubiquitin G76C and the E3 HECT ligase Huwe1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1, Polyubiquitin-B, SULFATE ION, ... | | Authors: | Jaeckl, M, Hartmann, M.D, Wiesner, S. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | beta-Sheet Augmentation Is a Conserved Mechanism of Priming HECT E3 Ligases for Ubiquitin Ligation.
J. Mol. Biol., 430, 2018
|
|
6FMJ
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-Acetamidopropanethioyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl) benzyl)pyrrolidine-2-carboxamide (ligand 3) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-acetamidopropanethioyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Soares, P, Lucas, X, Ciulli, A. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Thioamide substitution to probe the hydroxyproline recognition of VHL ligands.
Bioorg. Med. Chem., 26, 2018
|
|
