1RHH
 
 | | Crystal Structure of the Broadly HIV-1 Neutralizing Fab X5 at 1.90 Angstrom Resolution | | Descriptor: | Fab X5, heavy chain, light chain | | Authors: | Darbha, R, Phogat, S, Labrijn, A.F, Shu, Y, Gu, Y, Andrykovitch, M, Zhang, M.Y, Pantophlet, R, Martin, L, Vita, C, Burton, D.R, Dimitrov, D.S, Ji, X. | | Deposit date: | 2003-11-14 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Broadly Cross-Reactive HIV-1-Neutralizing Fab X5 and Fine Mapping of Its Epitope
Biochemistry, 43, 2004
|
|
3JCX
 
 | | Canine Parvovirus complexed with Fab E | | Descriptor: | Capsid protein 2, Fab E heavy chain, Fab E light chain | | Authors: | Organtini, L.J, Iketani, S, Huang, K, Ashley, R.E, Makhov, A.M, Conway, J.F, Parrish, C.R, Hafenstein, S. | | Deposit date: | 2016-03-21 | | Release date: | 2016-07-20 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Near-Atomic Resolution Structure of a Highly Neutralizing Fab Bound to Canine Parvovirus.
J.Virol., 90, 2016
|
|
6PXH
 
 | | Crystal Structure of MERS-CoV S1-NTD bound with G2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIHYDROFOLIC ACID, ... | | Authors: | Wang, N, McLellan, J.S. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD.
Cell Rep, 28, 2019
|
|
3RZW
 
 | |
6EQC
 
 | | Cryo-EM reconstruction of a complex of a binding protein and human adenovirus C5 hexon | | Descriptor: | Hexon protein, scFv of 9C12 antibody | | Authors: | Schmid, M, Ernst, P, Honegger, A, Suomalainen, M, Zimmermann, M, Braun, L, Stauffer, S, Thom, C, Dreier, B, Eibauer, M, Kipar, A, Vogel, V, Greber, U.F, Medalia, O, Plueckthun, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-02-07 | | Last modified: | 2018-02-14 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Adenoviral vector with shield and adapter increases tumor specificity and escapes liver and immune control.
Nat Commun, 9, 2018
|
|
3UYO
 
 | |
3BN9
 
 | | Crystal Structure of MT-SP1 in complex with Fab Inhibitor E2 | | Descriptor: | 1,2-ETHANEDIOL, E2 Fab Heavy Chain, E2 Fab Light Chain, ... | | Authors: | Farady, C.J, Schneider, E.L, Egea, P.F, Goetz, D.H, Craik, C.S. | | Deposit date: | 2007-12-13 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | Structure of an Fab-protease complex reveals a highly specific non-canonical mechanism of inhibition
J.Mol.Biol., 380, 2008
|
|
3MXV
 
 | |
3MXW
 
 | |
1ETZ
 
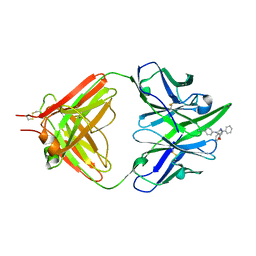 | | THE THREE-DIMENSIONAL STRUCTURE OF AN ANTI-SWEETENER FAB, NC10.14, SHOWS THE EXTENT OF STRUCTURAL DIVERSITY IN ANTIGEN RECOGNITION BY IMMUNOGLOBULINS | | Descriptor: | FAB NC10.14 - HEAVY CHAIN, FAB NC10.14 - LIGHT CHAIN, N-(P-CYANOPHENYL)-N'-DIPHENYLMETHYL-GUANIDINE-ACETIC ACID | | Authors: | Guddat, L.W, Shan, L, Broomell, C, Ramsland, P.A, Fan, Z, Anchin, J.M, Linthicum, D.S, Edmundson, A.B. | | Deposit date: | 2000-04-13 | | Release date: | 2000-10-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The three-dimensional structure of a complex of a murine Fab (NC10. 14) with a potent sweetener (NC174): an illustration of structural diversity in antigen recognition by immunoglobulins.
J.Mol.Biol., 302, 2000
|
|
8V7O
 
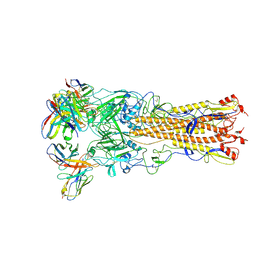 | |
1HZH
 
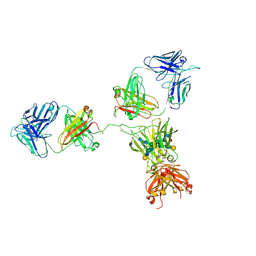 | | CRYSTAL STRUCTURE OF THE INTACT HUMAN IGG B12 WITH BROAD AND POTENT ACTIVITY AGAINST PRIMARY HIV-1 ISOLATES: A TEMPLATE FOR HIV VACCINE DESIGN | | Descriptor: | IMMUNOGLOBULIN HEAVY CHAIN, IMMUNOGLOBULIN LIGHT CHAIN,Uncharacterized protein, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saphire, E.O, Burton, D.R, Wilson, I.A. | | Deposit date: | 2001-01-24 | | Release date: | 2001-08-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design.
Science, 293, 2001
|
|
8DT8
 
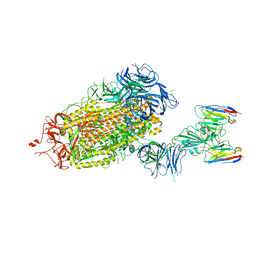 | | LM18/Nb136 bispecific tetra-nanobody immunoglobulin in complex with SARS-CoV-2-6P-Mut7 S protein (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LM18 nanobody, Nb136 nanobody, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Fully synthetic platform to rapidly generate tetravalent bispecific nanobody-based immunoglobulins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1MHH
 
 | |
6VKN
 
 | | BG505 SOSIP.v5.2.N241.N289 in complex with rhesus macaque Fab RM19R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-08-12 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Targeting HIV Env immunogens to B cell follicles in nonhuman primates through immune complex or protein nanoparticle formulations.
NPJ Vaccines, 5, 2020
|
|
2D03
 
 | | Crystal structure of the G91S mutant of the NNA7 Fab | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Xie, K, Song, S.C, Spitalnik, S.L, Wedekind, J.E. | | Deposit date: | 2005-07-23 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystallographic analysis of the NNA7 Fab and proposal for the mode of human blood-group recognition.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6QU9
 
 | | Fab fragment of an antibody that inhibits polymerisation of alpha-1-antitrypsin | | Descriptor: | FAB 4B12 heavy chain, FAB 4B12 light chain, GLYCEROL, ... | | Authors: | Jagger, A.M, Heyer-Chauhan, N, Lomas, D.A, Irving, J.A. | | Deposit date: | 2019-02-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for Z alpha 1 -antitrypsin polymerization in the liver.
Sci Adv, 6, 2020
|
|
5TRU
 
 | | Structure of the first-in-class checkpoint inhibitor Ipilimumab bound to human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, Ipilimumab Fab heavy chain, Ipilimumab Fab light chain | | Authors: | Ramagopal, U.A, Liu, W, Garrett-Thomson, S.C, Yan, Q, Srinivasan, M, Wong, S.C, Bell, A, Mankikar, S, Rangan, V.S, Deshpande, S, Bonanno, J.B, Korman, A.J, Almo, S.C. | | Deposit date: | 2016-10-27 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1PKQ
 
 | | Myelin Oligodendrocyte Glycoprotein-(8-18C5) Fab-complex | | Descriptor: | (8-18C5) chimeric Fab, heavy chain, light chain, ... | | Authors: | Breithaupt, C, Schubart, A, Zander, H, Skerra, A, Huber, R, Linington, C, Jacob, U. | | Deposit date: | 2003-06-06 | | Release date: | 2003-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the antigenicity of myelin oligodendrocyte glycoprotein
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
6Y54
 
 | | Crystal structure of a Neisseria meningitidis serogroup A capsular oligosaccharide bound to a functional Fab | | Descriptor: | 1,2-ETHANEDIOL, Fab A1.1 H chain, Fab A1.1 L chain, ... | | Authors: | Dello Iacono, L, Henriques, P, Adamo, R. | | Deposit date: | 2020-02-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structure of a protective epitope reveals the importance of acetylation of Neisseria meningitidis serogroup A capsular polysaccharide.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8EKF
 
 | |
8VUC
 
 | | Structure of FabS1CE2-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, S1CE2 VARIANT OF FAB-EPR-1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-29 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VUA
 
 | | Structure of FabS1CE1-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor | | Descriptor: | S1CE1 VARIANT OF FAB-EPR-1 heavy chain, S1CE1 VARIANT OF FAB-EPR-1 light chain | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-29 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
9ERX
 
 | | Structural basis of D9-THC analog activity at the Cannabinoid 1 receptor | | Descriptor: | (6aR,10aR)-9-(hydroxymethyl)-6,6-dimethyl-3-(2-methyloctan-2-yl)-6a,7,10,10a-tetrahydrobenzo[c]chromen-1-ol, Antibody ScFv16 Fab fragment, Cannabinoid receptor 1, ... | | Authors: | Thorsen, T.S, Kulkarni, Y, Boggild, A, Drace, T, Nissen, P, Gajhede, M, Boesen, T, Kastrup, J.S, Gloriam, D. | | Deposit date: | 2024-03-25 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of Delta 9 -THC analog activity at the Cannabinoid 1 receptor.
Res Sq, 2024
|
|
8VTR
 
 | | Structure of FabS1CE3-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor (orthorhombic form) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
