4M4Z
 
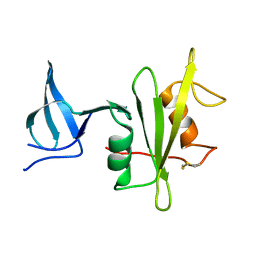 | |
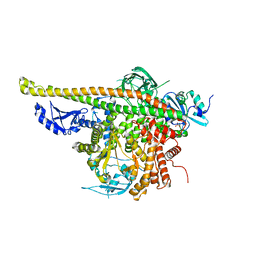
4L1B
 
 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, SULFATE ION | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-03 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
1XA6
 
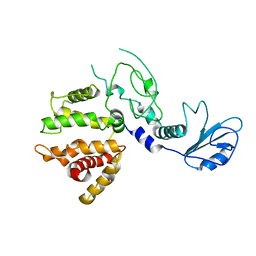 | | Crystal Structure of the Human Beta2-Chimaerin | | Descriptor: | Beta2-chimaerin, ZINC ION | | Authors: | Canagarajah, B, Leskow, F.C, Ho, J.Y, Mischak, H, Saidi, L.F, Kazanietz, M.G, Hurley, J.H. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mechanism for lipid activation of the Rac-specific GAP, beta2-chimaerin.
Cell(Cambridge,Mass.), 119, 2004
|
|
7YQE
 
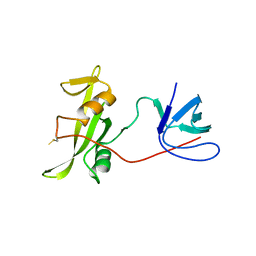 | |
4LUD
 
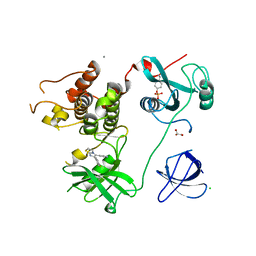 | | Crystal Structure of HCK in complex with the fluorescent compound SKF86002 | | Descriptor: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4L2Y
 
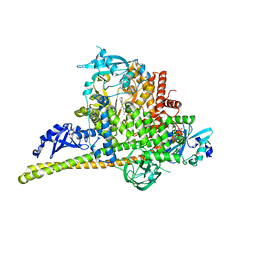 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha and compound 9d | | Descriptor: | 3-amino-5-[4-(morpholin-4-yl)pyrido[3',2':4,5]furo[3,2-d]pyrimidin-2-yl]phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
4LUE
 
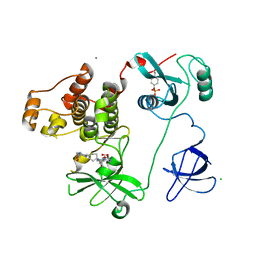 | | Crystal Structure of HCK in complex with 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (resulting from displacement of SKF86002) | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7ZLO
 
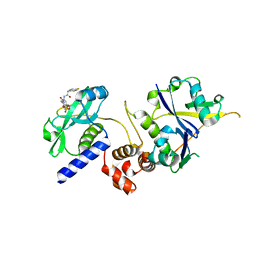 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 12 | | Descriptor: | Elongin-B, Elongin-C, Suppressor of cytokine signaling 2, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
7ZLP
 
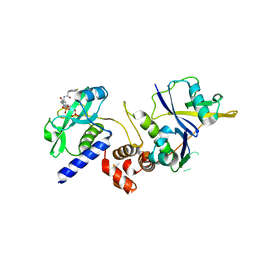 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 9 | | Descriptor: | Elongin-B, Elongin-C, PHOSPHATE ION, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
7ZLN
 
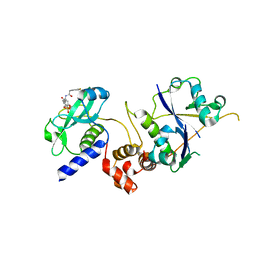 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 11 | | Descriptor: | Elongin-B, Elongin-C, Suppressor of cytokine signaling 2, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
7ZLS
 
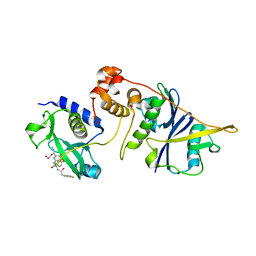 | | co-crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 13 | | Descriptor: | 1,2-ETHANEDIOL, Elongin-B, Elongin-C, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
7ZLR
 
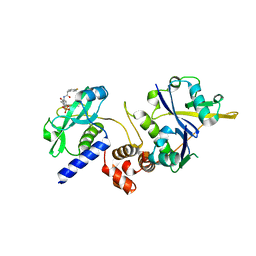 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 13 | | Descriptor: | Elongin-B, Elongin-C, Suppressor of cytokine signaling 2, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
7ZLM
 
 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound MN551 | | Descriptor: | Elongin-B, Elongin-C, Suppressor of cytokine signaling 2, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
4L23
 
 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha and PI-103 | | Descriptor: | 3-(4-MORPHOLIN-4-YLPYRIDO[3',2':4,5]FURO[3,2-D]PYRIMIDIN-2-YL)PHENOL, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-04 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
4K2R
 
 | | Structural basis for activation of ZAP-70 by phosphorylation of the SH2-kinase linker | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yan, Q, Barros, T, Visperas, P.R, Deindl, S, Kadlecek, T.A, Weiss, A, Kuriyan, J. | | Deposit date: | 2013-04-09 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Activation of ZAP-70 by Phosphorylation of the SH2-Kinase Linker.
Mol.Cell.Biol., 33, 2013
|
|
7ZN7
 
 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and rat STAT2 CCD | | Descriptor: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | Authors: | Lauer, S, Spahn, C.M.T, Schwefel, D. | | Deposit date: | 2022-04-20 | | Release date: | 2022-11-09 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
7ZNN
 
 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and full-length rat STAT2 | | Descriptor: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | Authors: | Lauer, S, Spahn, C.M.T, Schwefel, D. | | Deposit date: | 2022-04-21 | | Release date: | 2022-11-09 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
7Z3J
 
 | | Structure of crystallisable rat Phospholipase C gamma 1 in complex with inositol 1,4,5-trisphosphate | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Pinotsis, N, Bunney, T.D, Katan, M. | | Deposit date: | 2022-03-02 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the membrane interactions of phospholipase C gamma reveals key features of the active enzyme.
Sci Adv, 8, 2022
|
|
8AM0
 
 | | Crystal structure of human T1061E PI3Kalpha in complex with its regulatory subunit and the inhibitor GDC-0077 (Inavolisib) | | Descriptor: | (2R)-2-[[2-[(4S)-4-[bis(fluoranyl)methyl]-2-oxidanylidene-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl]amino]propanamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Goncalves, M, Johnson, J.L, Roewer, K.M. | | Deposit date: | 2022-08-02 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Epinephrine inhibits PI3K alpha via the Hippo kinases.
Cell Rep, 42, 2023
|
|
1Y57
 
 | | Structure of unphosphorylated c-Src in complex with an inhibitor | | Descriptor: | 4-[(4-METHYLPIPERAZIN-1-YL)METHYL]-N-{3-[(4-PYRIDIN-3-YLPYRIMIDIN-2-YL)AMINO]PHENYL}BENZAMIDE, Proto-oncogene tyrosine-protein kinase Src, SULFATE ION | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Manley, P.W, Jahnke, W, Fabbro, D, Liebetanz, J, Meyer, T. | | Deposit date: | 2004-12-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Crystal Structure of a c-Src Complex in an Active Conformation Suggests Possible Steps in c-Src Activation
Structure, 13, 2005
|
|
2ABL
 
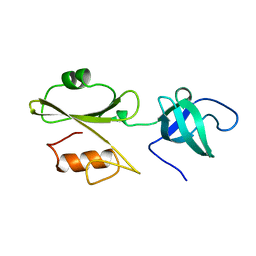 | |
1Y1U
 
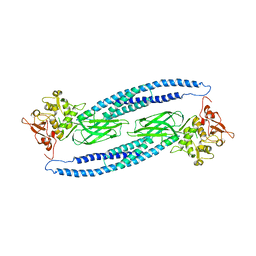 | | Structure of unphosphorylated STAT5a | | Descriptor: | Signal transducer and activator of transcription 5A | | Authors: | Neculai, D, Neculai, A.M, Verrier, S, Straub, K, Klumpp, K, Pfitzner, E, Becker, S. | | Deposit date: | 2004-11-19 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structure of the unphosphorylated STAT5a dimer
J.Biol.Chem., 280, 2005
|
|
1YVL
 
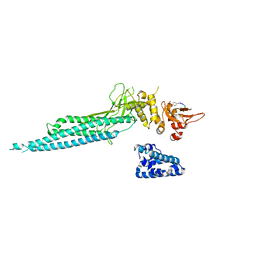 | | Structure of Unphosphorylated STAT1 | | Descriptor: | 5-residue peptide, GOLD ION, Signal transducer and activator of transcription 1-alpha/beta | | Authors: | Mao, X, Ren, Z, Parker, G.N, Sondermann, H, Pastorello, M.A, Wang, W, McMurray, J.S, Demeler, B, Darnell Jr, J.E, Chen, X. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural bases of unphosphorylated STAT1 association and receptor binding.
Mol.Cell, 17, 2005
|
|
4NWF
 
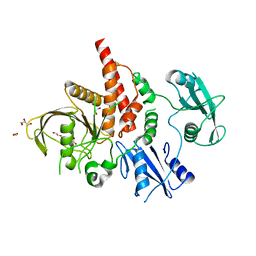 | | Crystal structure of the tyrosine phosphatase SHP-2 with N308D mutation | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battalie, K, Chirgadze, N.Y. | | Deposit date: | 2013-12-06 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with N308D mutation
To be Published
|
|
4NWG
 
 | | Crystal structure of the tyrosine phosphatase SHP-2 with E139D mutation | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battalie, K, Chirgadze, N.Y. | | Deposit date: | 2013-12-06 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with E139D mutation
To be Published
|
|
