1QJ1
 
 | | Novel Covalent Active Site Thrombin Inhibitors | | Descriptor: | 6-CARBAMIMIDOYL-2-[2-HYDROXY-6-(4-HYDROXY-PHENYL)-INDAN-1-YL]-HEXANOIC ACID, HIRUGEN, THROMBIN | | Authors: | Jhoti, H, Cleasby, A. | | Deposit date: | 1999-06-21 | | Release date: | 2000-06-22 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Thrombin Complexed to a Novel Series of Synthetic Inhibitors Containing a 5,5-Trans-Lactone Template
Biochemistry, 38, 1999
|
|
1QA7
 
 | | CRYSTAL COMPLEX OF THE 3C PROTEINASE FROM HEPATITIS A VIRUS WITH ITS INHIBITOR AND IMPLICATIONS FOR THE POLYPROTEIN PROCESSING IN HAV | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, HAV 3C PROTEINASE, ... | | Authors: | Bergmann, E.M, Cherney, M.M, Mckendrick, J, Vederas, J.C, James, M.N.G. | | Deposit date: | 1999-04-15 | | Release date: | 1999-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an inhibitor complex of the 3C proteinase from hepatitis A virus (HAV) and implications for the polyprotein processing in HAV.
Virology, 265, 1999
|
|
1QTF
 
 | | CRYSTAL STRUCTURE OF EXFOLIATIVE TOXIN B | | Descriptor: | EXFOLIATIVE TOXIN B | | Authors: | Vath, G.M, Earhart, C.A, Monie, D.D, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1999-06-27 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of exfoliative toxin B: a superantigen with enzymatic activity.
Biochemistry, 38, 1999
|
|
4QLQ
 
 | | yCP in complex with tripeptidic epoxyketone inhibitor 8 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-(morpholin-4-ylacetyl)-L-alanyl-N-[(2S,4R)-1-cyclohexyl-5-hydroxy-4-methyl-3-oxopentan-2-yl]-O-methyl-L-tyrosinamide, ... | | Authors: | De Bruin, G, Huber, E, Xin, B, Van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, Van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
4Q7X
 
 | | Neutrophil serine protease 4 (PRSS57) apo form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Serine protease 57, ... | | Authors: | Eigenbrot, C, Lin, S.J, Dong, K.C. | | Deposit date: | 2014-04-25 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of neutrophil serine protease 4 reveal an unusual mechanism of substrate recognition by a trypsin-fold protease.
Structure, 22, 2014
|
|
4QLT
 
 | | yCP in complex with tripeptidic epoxyketone inhibitor 2 (PR924) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-[(3-methyl-1H-inden-2-yl)carbonyl]-D-alanyl-N-[(2S,4R)-5-hydroxy-4-methyl-3-oxo-1-phenylpentan-2-yl]-L-tryptophanamide, ... | | Authors: | de Bruin, G, Huber, E, Xin, B, van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
3QZR
 
 | | Human enterovirus 71 3C protease mutant E71A in complex with rupintrivir | | Descriptor: | 1,2-ETHANEDIOL, 3C protein, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Wang, J, Fan, T, Yao, X, Wu, Z, Guo, L, Lei, X, Wang, J, Wang, M, Jin, Q, Cui, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.039 Å) | | Cite: | Crystal Structures of Enterovirus 71 3C Protease Complexed with Rupintrivir Reveal the Roles of Catalytically Important Residues.
J.Virol., 85, 2011
|
|
1QKI
 
 | | X-RAY STRUCTURE OF HUMAN GLUCOSE 6-PHOSPHATE DEHYDROGENASE (VARIANT CANTON R459L) COMPLEXED WITH STRUCTURAL NADP+ | | Descriptor: | GLUCOSE-6-PHOSPHATE 1-DEHYDROGENASE, GLYCEROL, GLYCOLIC ACID, ... | | Authors: | Au, S.W.N, Gover, S, Lam, V.M.S, Adams, M.J. | | Deposit date: | 1999-07-20 | | Release date: | 2000-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human Glucose-6-Phosphate Dehydrogenase: The Crystal Structure Reveals a Structural Nadp+ Molecule and Provides Insights Into Enzyme Deficiency
Structure, 8, 2000
|
|
1QL8
 
 | |
1QS8
 
 | | Crystal structure of the P. vivax aspartic proteinase plasmepsin complexed with the inhibitor pepstatin A | | Descriptor: | ACETATE ION, PEPSTATIN A, PLASMEPSIN | | Authors: | Khazanovich Bernstein, N, Cherney, M.M, Yowell, C.A, Dame, J.B, James, M.N.G. | | Deposit date: | 1999-06-25 | | Release date: | 1999-07-07 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation of P. vivax plasmepsin.
J.Mol.Biol., 329, 2003
|
|
3S9A
 
 | |
3SJ9
 
 | | crystal structure of the C147A mutant 3C of CVA16 in complex with FAGLRQAVTQ peptide | | Descriptor: | 3C protease, FAGLRQAVTQ peptide | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
1QL9
 
 | |
3SHO
 
 | |
3V6M
 
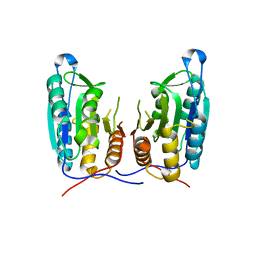 | | Inhibition of caspase-6 activity by single mutation outside the active site | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.J, Liu, D.F, Li, L.F, Su, X.D. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays
J.Biol.Chem., 287, 2012
|
|
3FKT
 
 | |
1QQU
 
 | |
3TK9
 
 | | Crystal structure of human granzyme H | | Descriptor: | Granzyme H, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
3H0B
 
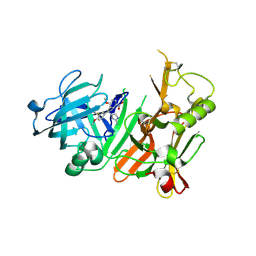 | | Discovery of aminoheterocycles as a novel beta-secretase inhibitor class | | Descriptor: | 4-[(1S)-1-(3-fluoro-4-methoxyphenyl)-2-(2-methoxy-5-nitrophenyl)ethyl]-1H-imidazol-2-amine, Beta-secretase 1 | | Authors: | Allison, T.J. | | Deposit date: | 2009-04-08 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of aminoheterocycles as a novel beta-secretase inhibitor class: pH dependence on binding activity part 1.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3RUO
 
 | | Complex structure of HevB EV93 main protease 3C with Rupintrivir (AG7088) | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, CHLORIDE ION, HEVB EV93 3C PROTEASE, ... | | Authors: | Kaczmarska, Z, Janowski, R, Costenaro, L, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2011-05-05 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
4PMJ
 
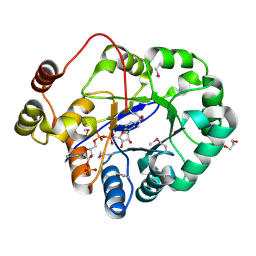 | | Crystal structure of a putative oxidoreductase from Sinorhizobium meliloti 1021 in complex with NADP | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Szlachta, K, Zimmerman, M.D, Hillerich, B.S, Gizzi, A, Toro, R, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-05-21 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative oxidoreductase from Sinorhizobiummeliloti 1021 in complex with NADP
to be published
|
|
1BDA
 
 | |
4PQH
 
 | | Crystal structure of glutathione transferase lambda1 from Populus trichocarpa | | Descriptor: | GLUTATHIONE, SODIUM ION, glutathione transferase lambda1 | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Prosper, P, Didierjean, C, Haouz, A, Saul, F, Rouhier, N, Hecker, A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and enzymatic insights into Lambda glutathione transferases from Populus trichocarpa, monomeric enzymes constituting an early divergent class specific to terrestrial plants.
Biochem.J., 462, 2014
|
|
4AGK
 
 | |
3EE0
 
 | | Crystal Structure of the W215A/E217A Mutant of Human Thrombin (space group P2(1)2(1)2(1)) | | Descriptor: | Thrombin heavy chain, Thrombin light chain | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-07-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular Basis for the Kinetic Differences of the W215A/E217A Mutant of Human and Murine Thrombin
To be Published
|
|
