6UOZ
 
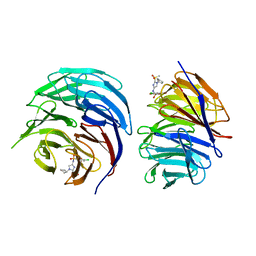 | |
4A8S
 
 | | Non-Catalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial dsRNA virus phi6 from De Novo Initiation to Elongation | | Descriptor: | 5'-D(*TP*TP*TP*TP*CP*GP*CP*GP*TP*AP*GP*CP*GP)-3', ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wright, S, Poranen, M.M, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Noncatalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial Double-Stranded RNA Virus Phi6 from De Novo Initiation to Elongation.
J.Virol., 86, 2012
|
|
1ZTR
 
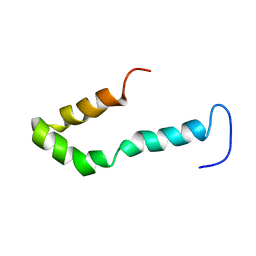 | | Solution structure of Engrailed homeodomain L16A mutant | | Descriptor: | Segmentation polarity homeobox protein engrailed | | Authors: | Religa, T.L, Markson, J.S, Mayor, U, Freund, S.M.V, Fersht, A.R. | | Deposit date: | 2005-05-27 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a protein denatured state and folding intermediate.
Nature, 437, 2005
|
|
6XQD
 
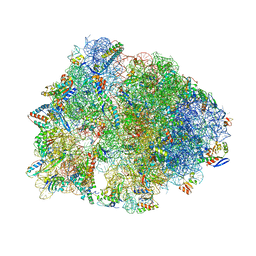 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with sarecycline, UUC-mRNA, and deacylated P-site tRNA at 2.80A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Batool, Z, Lomakin, I.B, Bunick, C.G, Polikanov, Y.S. | | Deposit date: | 2020-07-09 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Sarecycline interferes with tRNA accommodation and tethers mRNA to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
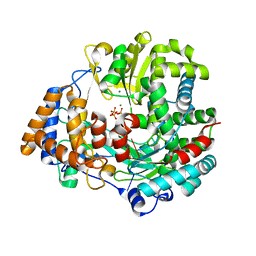
4YTR
 
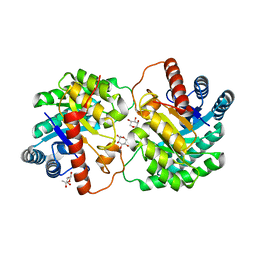 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy L-tagatose | | Descriptor: | 1-deoxy-L-tagatose, 1-deoxy-beta-L-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
6XHW
 
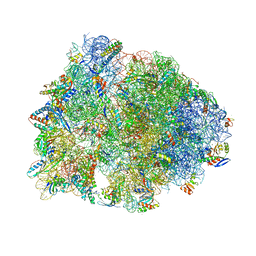 | | Crystal structure of the A2058-unmethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A- and P-site tRNAs, and deacylated E-site tRNA at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Syroegin, E.A, Aleksandrova, E.V, Atkinson, G.C, Gregory, S.T, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance.
Nat.Chem.Biol., 17, 2021
|
|
5FTJ
 
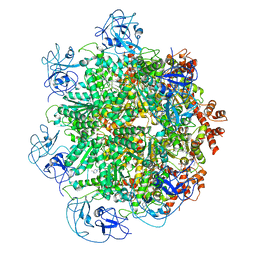 | | Cryo-EM structure of human p97 bound to UPCDC30245 inhibitor | | Descriptor: | 1-(3-(5-FLUORO-1H-INDOL-2-YL)PHENYL)PIPERIDIN-4-YL)(2-(4-ISOPROPYL-PIPERAZIN1-YL)ETHYL)-CARBAMATE, ADENOSINE-5'-DIPHOSPHATE, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | Authors: | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
6Q6H
 
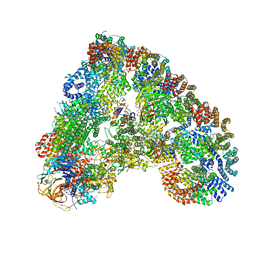 | |
4YTQ
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy D-tagatose | | Descriptor: | 1-deoxy-D-tagatose, 1-deoxy-alpha-D-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
5FHX
 
 | | CRYSTAL STRUCTURE OF CODV IN COMPLEX WITH IL4 AT 2.55 Ang. RESOLUTION. | | Descriptor: | Antibody fragment light chain, Interleukin-4, PHOSPHATE ION, ... | | Authors: | Vallee, F, Dupuy, A, Rak, A. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | CODV-Ig, a universal bispecific tetravalent and multifunctional immunoglobulin format for medical applications.
Mabs, 8, 2016
|
|
8VSP
 
 | | Cryo-EM structure of human invariant chain in complex with HLA-DQ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, ... | | Authors: | Wang, N, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural insights into human MHC-II association with invariant chain.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VRW
 
 | | Cryo-EM structure of human invariant chain in complex with HLA-DR15 | | Descriptor: | HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Wang, N, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2024-01-22 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights into human MHC-II association with invariant chain.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7RQ8
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
4YK8
 
 | |
7RQC
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MFI-tripeptidyl-tRNA analog ACCA-IFM at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8EZ0
 
 | | Cryo-EM structure of 4 insulins bound full-length mouse IR mutant with physically decoupled alpha CTs (C684S/C685S/C687S; denoted as IR-3CS) Symmetric conformation | | Descriptor: | Insulin, Insulin receptor | | Authors: | Li, J, Wu, J.Y, Hall, C, Bai, X.C, Choi, E. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for the role of disulfide-linked alpha CTs in the activation of insulin-like growth factor 1 receptor and insulin receptor.
Elife, 11, 2022
|
|
4YTS
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy 3-keto D-galactitol | | Descriptor: | 1-deoxy-D-xylo-hex-3-ulose, 1-deoxy-alpha-D-xylo-hex-3-ulofuranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
8EYX
 
 | | Cryo-EM structure of 4 insulins bound full-length mouse IR mutant with physically decoupled alpha CTs (C684S/C685S/C687S; denoted as IR-3CS) Asymmetric conformation 1 | | Descriptor: | Insulin, Insulin receptor | | Authors: | Li, J, Wu, J.Y, Hall, C, Bai, X.C, Choi, E. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-09 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular basis for the role of disulfide-linked alpha CTs in the activation of insulin-like growth factor 1 receptor and insulin receptor.
Elife, 11, 2022
|
|
7RQB
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MAI-tripeptidyl-tRNA analog ACCA-IAM at 2.45A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8EYY
 
 | | Cryo-EM structure of 4 insulins bound full-length mouse IR mutant with physically decoupled alpha CTs (C684S/C685S/C687S, denoted as IR-3CS) Asymmetric conformation 2 | | Descriptor: | Insulin, Insulin receptor | | Authors: | Li, J, Wu, J.Y, Hall, C, Bai, X.C, Choi, E. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-09 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Molecular basis for the role of disulfide-linked alpha CTs in the activation of insulin-like growth factor 1 receptor and insulin receptor.
Elife, 11, 2022
|
|
7RQ9
 
 | | Crystal structure of the A2058-dimethylated Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
5HBM
 
 | | Crystal Structure of a Dihydroxycoumarin RNase H Active-Site Inhibitor in Complex with HIV-1 Reverse Transcriptase | | Descriptor: | (7,8-dihydroxy-2-oxo-2H-chromen-4-yl)acetic acid, 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MANGANESE (II) ION, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2015-12-31 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.043 Å) | | Cite: | Crystal Structure of a Dihydroxycoumarin RNase H Active-Site Inhibitor in Complex with HIV-1 Reverse Transcriptase
To Be Published
|
|
4YTU
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with L-erythrulose | | Descriptor: | D-tagatose 3-epimerase, L-Erythrulose, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
8DTL
 
 | | Cryo-EM structure of insulin receptor (IR) bound with S597 peptide | | Descriptor: | Insulin mimetic peptide S597, Insulin receptor | | Authors: | Park, J, Li, J, Mayer, J.P, Ball, K.A, Wu, J.Y, Hall, C, Accili, D, Stowell, M.H.B, Bai, X.C, Choi, E. | | Deposit date: | 2022-07-25 | | Release date: | 2022-09-07 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Activation of the insulin receptor by an insulin mimetic peptide.
Nat Commun, 13, 2022
|
|
4YWU
 
 | | Structural insight into the substrate inhibition mechanism of NADP+-dependent succinic semialdehyde dehydrogenase from Streptococcus pyogenes | | Descriptor: | 4-oxobutanoic acid, SULFATE ION, Succinic semialdehyde dehydrogenase | | Authors: | Jang, E.H, Park, S.A, Chi, Y.M, Lee, K.S. | | Deposit date: | 2015-03-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into the substrate inhibition mechanism of NADP(+)-dependent succinic semialdehyde dehydrogenase from Streptococcus pyogenes.
Biochem.Biophys.Res.Commun., 461, 2015
|
|
