6UGF
 
 | |
7RSY
 
 | | HIV-1 gp120 complex with CJF-III-049-R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 Clade C1086, N~1~-{(1R,2R,3S)-2-(carbamimidamidomethyl)-3-[(3R)-3,4-dihydroxybutyl]-5-[(methylamino)methyl]-2,3-dihydro-1H-inden-1-yl}-N~2~-(4-chloro-3-fluorophenyl)ethanediamide | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of gp120 Residue His105 as a Novel Target for HIV-1 Neutralization by Small-Molecule CD4-Mimics.
Acs Med.Chem.Lett., 12, 2021
|
|
6WB0
 
 | | +3 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state) | | Descriptor: | HIV-1 viral RNA genome fragment, Reverse transcriptase/ribonuclease H, reverse transcriptase p51 subunit, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
7RSZ
 
 | | HIV-1 gp120 complex with CJF-II-204 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 Clade C1086, N~1~-{(1R,2R,3S)-2-(carbamimidamidomethyl)-3-[(3R)-3,4-dihydroxybutyl]-5-[(methylamino)methyl]-2,3-dihydro-1H-inden-1-yl}-N~2~-(4-chloro-3-fluorophenyl)ethanediamide, ... | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Identification of gp120 Residue His105 as a Novel Target for HIV-1 Neutralization by Small-Molecule CD4-Mimics.
Acs Med.Chem.Lett., 12, 2021
|
|
6VSA
 
 | |
7VH9
 
 | | Solution structure of the chimeric peptide of the first SURP domain of Human SF3A1 and the interacting region of SF1. | | Descriptor: | Splicing factor 3A subunit 1,Splicing factor 1 | | Authors: | Muto, Y, Kuwasako, K, Takizawa, M, Kobayashi, N, Sakamoto, T. | | Deposit date: | 2021-09-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction between the first SURP domain of the SF3A1 subunit in U2 snRNP and the human splicing factor SF1.
Protein Sci., 31, 2022
|
|
6UQK
 
 | | Cryo-EM structure of type 3 IP3 receptor revealing presence of a self-binding peptide | | Descriptor: | ZINC ION, inositol 1,4,5-triphosphate receptor, type 3 | | Authors: | Azumaya, C.M, Linton, E.A, Risener, C.J, Nakagawa, T, Karakas, E. | | Deposit date: | 2019-10-20 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Cryo-EM structure of human type-3 inositol triphosphate receptor reveals the presence of a self-binding peptide that acts as an antagonist.
J.Biol.Chem., 295, 2020
|
|
7XO2
 
 | |
7XO0
 
 | |
7XO3
 
 | |
7XO1
 
 | |
7VCT
 
 | | Human p97 single hexamer conformer III with D1-ATPgammaS and D2-ADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCX
 
 | | Human p97 single hexamer conformer II with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
6VYI
 
 | | Cryo-EM structure of human diacylglycerol O-acyltransferase 1 | | Descriptor: | Diacylglycerol O-acyltransferase 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Sui, X, Wang, K, Gluchowski, N, Liao, M, Walther, C.T, Farese, V.R. | | Deposit date: | 2020-02-26 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme.
Nature, 581, 2020
|
|
7VCS
 
 | | Human p97 double hexamer conformer II with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCV
 
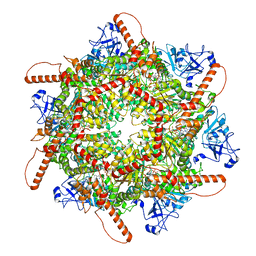 | | Human p97 single hexamer conformer I with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
6VQU
 
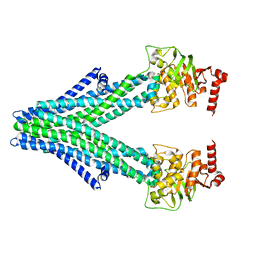 | | Structure of a bacterial Atm1-family ABC exporter | | Descriptor: | ATM1-type heavy metal exporter | | Authors: | Fan, C, Rees, D.C. | | Deposit date: | 2020-02-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | A structural framework for unidirectional transport by a bacterial ABC exporter.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WAZ
 
 | | +1 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state) | | Descriptor: | HIV-1 viral RNA genome fragment, Reverse transcriptase p51 subunit, Reverse transcriptase/ribonuclease H, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
6WB1
 
 | | +3 extended HIV-1 reverse transcriptase initiation complex core (intermediate state) | | Descriptor: | HIV-1 viral RNA genome fragment, REVERSE TRANSCRIPTASE/RIBONUCLEASE H, reverse transcriptase p51 subunit, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
7VKV
 
 | |
3C2K
 
 | | DNA POLYMERASE BETA with a gapped DNA substrate and DUMPNPP with Manganese in the active site | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*DCP*DCP*DGP*DAP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DCP*DAP*DGP*DC)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation.
Mol.Cell, 30, 2008
|
|
6VQR
 
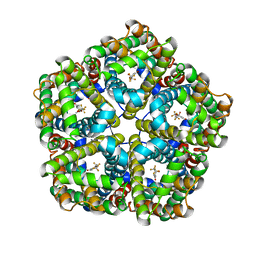 | | CryoEM Structure of the PfFNT-inhibitor complex | | Descriptor: | (2R)-2-hydroxy-7-methoxy-2-(pentafluoroethyl)-2,3-dihydro-4H-1-benzopyran-4-one, Formate-nitrite transporter | | Authors: | Su, C.C, Lyu, M. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural basis of transport and inhibition of the Plasmodium falciparum transporter PfFNT.
Embo Rep., 22, 2021
|
|
6W4F
 
 | | NMR-driven structure of KRAS4B-GDP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3C2M
 
 | | Ternary complex of DNA POLYMERASE BETA with a G:dAPCPP mismatch in the active site | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]adenosine, DNA (5'-D(*DCP*DCP*DGP*DAP*DCP*DGP*DGP*DCP*DGP*DCP*DAP*DTP*DCP*DAP*DGP*DC)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation.
Mol.Cell, 30, 2008
|
|
6VQT
 
 | |
