6OLZ
 
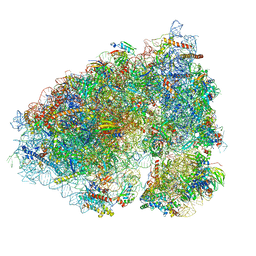 | |
7NRC
 
 | | Structure of the yeast Gcn1 bound to a leading stalled 80S ribosome with Rbg2, Gir2, A- and P-tRNA and eIF5A | | Descriptor: | 18S rRNA (1771-MER), 25S rRNA (3184-MER), 40S ribosomal protein S0-A, ... | | Authors: | Pochopien, A.A, Beckert, B, Wilson, D.N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of Gcn1 bound to stalled and colliding 80S ribosomes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
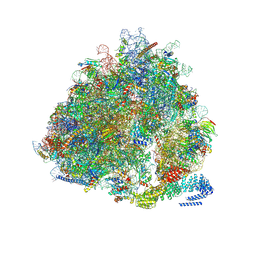
5A2Q
 
 | | Structure of the HCV IRES bound to the human ribosome | | Descriptor: | 18S RRNA, HCV IRES, MAGNESIUM ION, ... | | Authors: | Quade, N, Leiundgut, M, Boehringer, D, Heuvel, J.v.d, Ban, N. | | Deposit date: | 2015-05-21 | | Release date: | 2015-07-15 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-Em Structure of Hepatitis C Virus Ires Bound to the Human Ribosome at 3.9 Angstrom Resolution
Nat.Commun., 6, 2015
|
|
3J7R
 
 | | Structure of the translating mammalian ribosome-Sec61 complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
7KAS
 
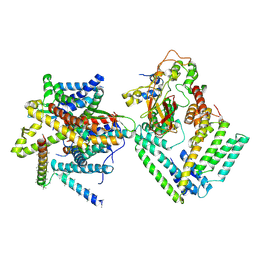 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec63 FN3 mutant, class with Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6R7Q
 
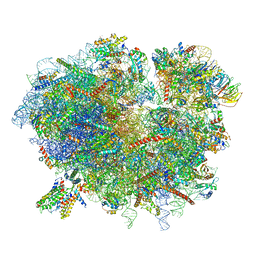 | | Structure of XBP1u-paused ribosome nascent chain complex with Sec61. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shanmuganathan, V, Cheng, J, Braunger, K, Berninghausen, O, Beatrix, B, Beckmann, R. | | Deposit date: | 2019-03-29 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and mutational analysis of the ribosome-arresting human XBP1u.
Elife, 8, 2019
|
|
8BQD
 
 | | Yeast 80S ribosome in complex with Map1 (conformation 1) | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
6ZQB
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state B2 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
7SYV
 
 | | Structure of the wt IRES eIF5B-containing pre-48S initiation complex, open conformation. Structure 14(wt) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-20 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
3JBM
 
 | | Electron cryo-microscopy of a virus-like particle of orange-spotted grouper nervous necrosis virus | | Descriptor: | virus-like particle of orange-spotted grouper nervous necrosis virus | | Authors: | Xie, J, Li, K, Gao, Y, Huang, R, Lai, Y, Shi, Y, Yang, S, Zhu, G, Zhang, Q, He, J. | | Deposit date: | 2015-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis and insertion study reveal the ideal sites for surface displaying foreign peptides on a betanodavirus-like particle
Vet. Res., 47, 2016
|
|
5LL6
 
 | | Structure of the 40S ABCE1 post-splitting complex in ribosome recycling and translation initiation | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Heuer, A, Gerovac, M, Schmidt, C, Trowitzsch, S, Preis, A, Koetter, P, Berninghausen, O, Becker, T, Beckmann, R, Tampe, R. | | Deposit date: | 2016-07-26 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the 40S-ABCE1 post-splitting complex in ribosome recycling and translation initiation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5FLX
 
 | | Mammalian 40S HCV-IRES complex | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Yamamoto, H, Collier, M, Loerke, J, Ismer, J, Schmidt, A, Hilal, T, Sprink, T, Yamamoto, K, Mielke, T, Burger, J, Shaikh, T.R, Dabrowski, M, Hildebrand, P.W, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Architecture of the Ribosome-Bound Hepatitis C Virus Internal Ribosomal Entry Site RNA.
Embo J., 34, 2015
|
|
6HCF
 
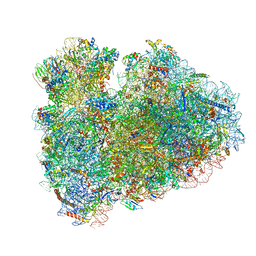 | | Structure of the rabbit 80S ribosome stalled on globin mRNA at the stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6BQN
 
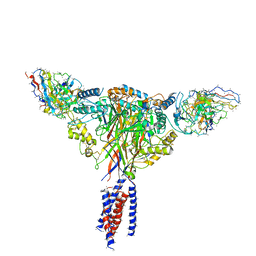 | | Cryo-EM structure of ENaC | | Descriptor: | 10D4 fab, 7B1 fab, EGFP-SCNN1G chimera, ... | | Authors: | Noreng, S, Bharadwaj, A, Posert, R, Yoshioka, C, Baconguis, I. | | Deposit date: | 2017-11-28 | | Release date: | 2018-10-10 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the human epithelial sodium channel by cryo-electron microscopy.
Elife, 7, 2018
|
|
6GQB
 
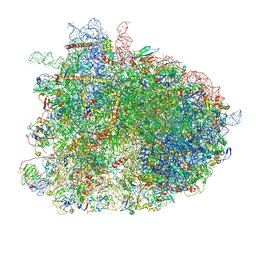 | | Cryo-EM reconstruction of yeast 80S ribosome in complex with mRNA, tRNA and eEF2 (GDP+AlF4/sordarin) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Pellegrino, S, Yusupov, M, Yusupova, G, Hashem, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-07-11 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insights into the Role of Diphthamide on Elongation Factor 2 in mRNA Reading-Frame Maintenance.
J. Mol. Biol., 430, 2018
|
|
7V80
 
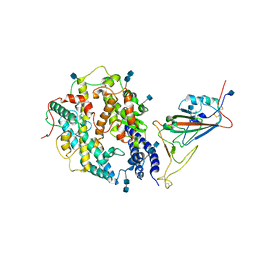 | | Local refinement of SARS-CoV-2 S-Beta variant (B.1.351) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Local refinement of SARS-CoV-2 S-Beta variant (B.1.351) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain
To Be Published
|
|
4V5O
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC 40S RIBOSOMAL SUBUNIT IN COMPLEX WITH INITIATION FACTOR 1. | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S12, 40S RIBOSOMAL PROTEIN S3A, ... | | Authors: | Rabl, J, Leibundgut, M, Ataide, S.F, Haag, A, Ban, N. | | Deposit date: | 2010-11-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1.
Science, 331, 2011
|
|
3QLV
 
 | |
4W69
 
 | |
5LZY
 
 | | Structure of the mammalian rescue complex with Pelota and Hbs1l assembled on a polyadenylated mRNA. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
7SUK
 
 | | Structure of Bfr2-Lcp5 Complex Observed in the Small Subunit Processome Isolated from R2TP-depleted Yeast Cells | | Descriptor: | 18S pre-rRNA, 40S ribosomal protein S11-A, 40S ribosomal protein S13, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-11-17 | | Release date: | 2022-07-06 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Artificial intelligence-assisted cryoEM structure of Bfr2-Lcp5 complex observed in the yeast small subunit processome.
Commun Biol, 5, 2022
|
|
5JUO
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure I (fully rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
6RXV
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B2 | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6GQV
 
 | | Cryo-EM recosntruction of yeast 80S ribosome in complex with mRNA, tRNA and eEF2 (GMPPCP) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Pellegrino, S, Yusupov, M, Yusupova, G, Hashem, Y. | | Deposit date: | 2018-06-08 | | Release date: | 2018-07-11 | | Last modified: | 2018-08-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Insights into the Role of Diphthamide on Elongation Factor 2 in mRNA Reading-Frame Maintenance.
J. Mol. Biol., 430, 2018
|
|
7KAO
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore mutant, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
