8TQM
 
 | |
4R3R
 
 | | Crystal structures of EGFR in complex with Mig6 | | Descriptor: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1' | | Authors: | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | Deposit date: | 2014-08-17 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4R3P
 
 | | Crystal structures of EGFR in complex with Mig6 | | Descriptor: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1 | | Authors: | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | Deposit date: | 2014-08-17 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6XMT
 
 | | Structure of P5A-ATPase Spf1, BeF3-bound form | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, P5A-type ATPase | | Authors: | Park, E, Sim, S.I. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase.
Science, 369, 2020
|
|
6XMQ
 
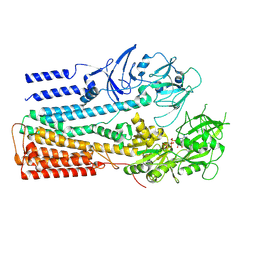 | | Structure of P5A-ATPase Spf1, AMP-PCP-bound form | | Descriptor: | MAGNESIUM ION, P5A-type ATPase, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Park, E, Sim, S.I. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase.
Science, 369, 2020
|
|
6XMU
 
 | |
6XMS
 
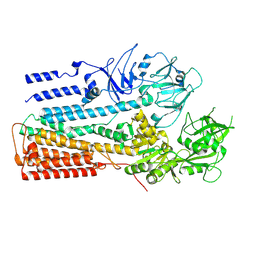 | | Structure of P5A-ATPase Spf1, AlF4-bound form | | Descriptor: | MAGNESIUM ION, P5A-type ATPase, TETRAFLUOROALUMINATE ION | | Authors: | Park, E, Sim, S.I. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase.
Science, 369, 2020
|
|
8DO3
 
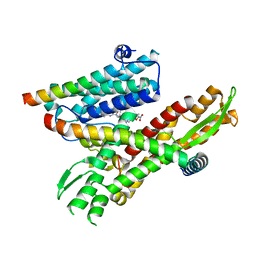 | | Cryo-EM structure of the human Sec61 complex inhibited by eeyarestatin I | | Descriptor: | N'-(4-chlorophenyl)-N-[(4R)-3-(4-chlorophenyl)-5,5-dimethyl-1-(2-{(2E)-2-[(2E)-3-(5-nitrofuran-2-yl)prop-2-en-1-ylidene]hydrazinyl}-2-oxoethyl)-2-oxoimidazolidin-4-yl]-N-hydroxyurea, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DNW
 
 | |
8DNY
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by decatransin | | Descriptor: | Decatransin peptide inhibitor, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DNX
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by cotransin | | Descriptor: | Cotransin analogue peptide inhibitor, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DO2
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by cyclotriazadisulfonamide (CADA) | | Descriptor: | 9-benzyl-1,5-bis(4-methylbenzene-1-sulfonyl)-3-methylidene-1,5,9-triazacyclododecane, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DNV
 
 | |
8DO0
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by mycolactone | | Descriptor: | Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, Protein transport protein Sec61 subunit gamma, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DO1
 
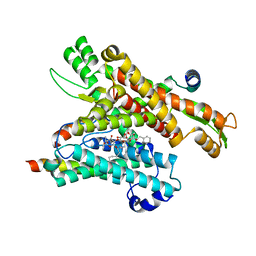 | | Cryo-EM structure of the human Sec61 complex inhibited by ipomoeassin F | | Descriptor: | Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, Protein transport protein Sec61 subunit gamma, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DNZ
 
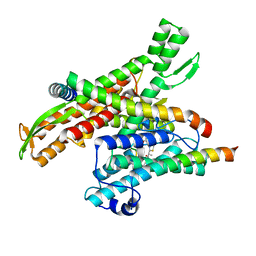 | | Cryo-EM structure of the human Sec61 complex inhibited by apratoxin F | | Descriptor: | Apratoxin F peptide inhibitor, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8QPP
 
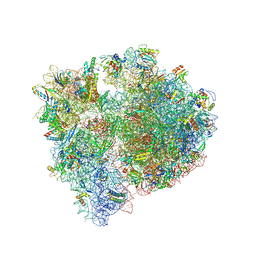 | | Bacillus subtilis MutS2-collided disome complex (stalled 70S) | | Descriptor: | 16S rRNA (1533-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Park, E, Mackens-Kiani, T, Berhane, R, Esser, H, Erdenebat, C, Burroughs, A.M, Berninghausen, O, Aravind, L, Beckmann, R, Green, R, Buskirk, A.R. | | Deposit date: | 2023-10-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | B. subtilis MutS2 splits stalled ribosomes into subunits without mRNA cleavage.
Embo J., 43, 2024
|
|
8R55
 
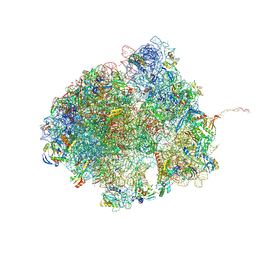 | | Bacillus subtilis MutS2-collided disome complex (collided 70S) | | Descriptor: | 16S rRNA (1533-MER), 23S RNA (2887-MER), 30S ribosomal protein S10, ... | | Authors: | Park, E, Mackens-Kiani, T, Berhane, R, Esser, H, Erdenebat, C, Burroughs, A.M, Berninghausen, O, Aravind, L, Beckmann, R, Green, R, Buskirk, A.R. | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | B. subtilis MutS2 splits stalled ribosomes into subunits without mRNA cleavage.
Embo J., 43, 2024
|
|
6COY
 
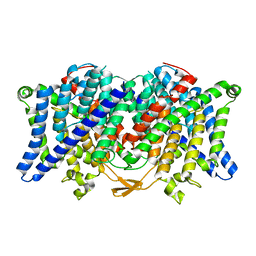 | |
6COZ
 
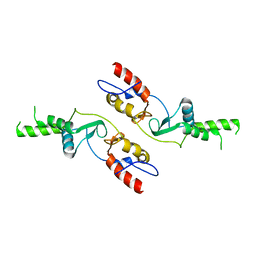 | |
6D8E
 
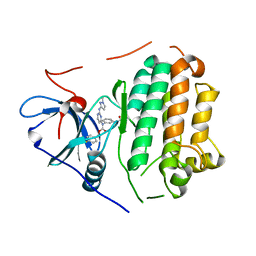 | |
6DUK
 
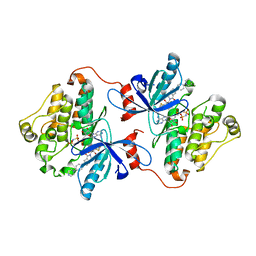 | | EGFR with an allosteric inhibitor | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-{1-oxo-6-[4-(piperazin-1-yl)phenyl]-1,3-dihydro-2H-isoindol-2-yl}-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Park, E, Eck, M.J. | | Deposit date: | 2018-06-21 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Single and Dual Targeting of Mutant EGFR with an Allosteric Inhibitor.
Cancer Discov, 9, 2019
|
|
6UCU
 
 | | Cryo-EM structure of the mitochondrial TOM complex from yeast (dimer) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Park, E, Tucker, K. | | Deposit date: | 2019-09-17 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6UCV
 
 | | Cryo-EM structure of the mitochondrial TOM complex from yeast (tetramer) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Park, E, Tucker, K. | | Deposit date: | 2019-09-17 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6XMP
 
 | | Structure of P5A-ATPase Spf1, Apo form | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, P5A-type ATPase | | Authors: | Park, E, Sim, S.I. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase.
Science, 369, 2020
|
|
