8WML
 
 | |
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8WT9
 
 | | Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction resolution) | | Descriptor: | IS621 transposase, MAGNESIUM ION, bridge RNA, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishimasu, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanism of bridge RNA-guided recombination.
Nature, 630, 2024
|
|
8XOP
 
 | |
8VE7
 
 | |
7S2S
 
 | | nanobody bound to Interleukin-2Rbeta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IL2Rb-binding nanobody, Interleukin-2 receptor subunit beta, ... | | Authors: | Glassman, C.R, Jude, K.M, Yen, M, Garcia, K.C. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Facile discovery of surrogate cytokine agonists.
Cell, 185, 2022
|
|
8VDZ
 
 | | A designed tetrahedral protein scaffold - DARP14 | | Descriptor: | Subunit A, Subunit B | | Authors: | Suder, D.S, Gonen, S. | | Deposit date: | 2023-12-18 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mitigating the Blurring Effect of CryoEM Averaging on a Flexible and Highly Symmetric Protein Complex through Sub-Particle Reconstruction.
Int J Mol Sci, 25, 2024
|
|
4YPM
 
 | |
8Z8N
 
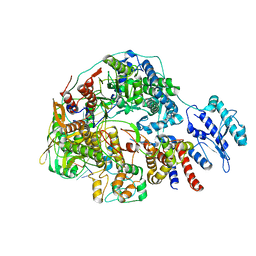 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 3 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8XBX
 
 | | The cryo-EM structure of the RAD51 L2 loop bound to the linker DNA with the blunt end of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8WMJ
 
 | | structure of PSI-11CAC complex at Logrithmic growth phase | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
8WU5
 
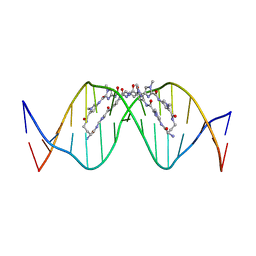 | | The complex of CAG repeat sequence-specific binding cPIP and dsDNA with A-A mismatch | | Descriptor: | (1^2Z,4^2Z,11^2Z,14^2Z,22^2Z,25^2Z,32^2Z,35^2Z,19R,40R)-1^1,4^1,11^1,14^1,22^1,25^1,32^1,35^1-octamethyl-2,5,9,12,15,20,23,26,30,33,36,41-dodecaoxo-1^1H,4^1H,11^1H,14^1H,22^1H,25^1H,32^1H,35^1H-3,6,10,13,16,21,24,27,31,34,37,42-dodecaaza-1(2,4),11,22,32(4,2)-tetraimidazola-4,14,25,35(4,2)-tetrapyrrolacyclodotetracontaphane-19,40-diaminium, DNA (5'-D(*GP*CP*(CBR)P*GP*AP*GP*CP*AP*GP*CP*AP*CP*GP*GP*C)-3') | | Authors: | Abe, K, Takeda, K, Sugiyama, H. | | Deposit date: | 2023-10-20 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of a Complex of a CAG/CTG Repeat Sequence-Specific Binding Molecule and A-A-Mismatch-Containing DNA.
Jacs Au, 4, 2024
|
|
7S2R
 
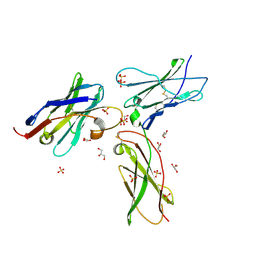 | | nanobody bound to IL-2Rg | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, ... | | Authors: | Glassman, C.R, Jude, K.M, Yen, M, Garcia, K.C. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Facile discovery of surrogate cytokine agonists.
Cell, 185, 2022
|
|
9CEQ
 
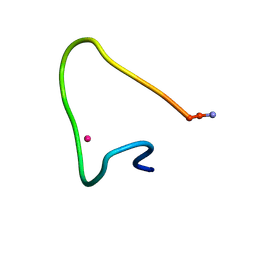 | |
8ZMR
 
 | | Vesamicol-bound VAChT | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Vesicular acetylcholine transporter,DARPinoff7, vesamicol | | Authors: | Zhang, Z, Zhang, Y, Dai, F, Zhang, Y.X, Lee, C.-H. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into VAChT neurotransmitter recognition and inhibition.
Cell Res., 2024
|
|
8X7A
 
 | | Treprostinil bound Prostacyclin Receptor G protein complex | | Descriptor: | 2-[[(1~{R},2~{R},3~{a}~{S},9~{a}~{S})-2-oxidanyl-1-[(3~{S})-3-oxidanyloctyl]-2,3,3~{a},4,9,9~{a}-hexahydro-1~{H}-cyclopenta[g]naphthalen-5-yl]oxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.J, Jin, S, Zhang, H, Xu, Y, Hu, W, Jiang, Y, Chen, C, Wang, D.W, Xu, H.E, Wu, C. | | Deposit date: | 2023-11-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Molecular recognition and activation of the prostacyclin receptor by anti-pulmonary arterial hypertension drugs.
Sci Adv, 10, 2024
|
|
9G2A
 
 | | Staphylococcus aureus MazF in complex with nanobody 4 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endoribonuclease MazF, ... | | Authors: | Prolic-Kalinsek, M, Zorzini, V, Haesaerts, S, Loris, R. | | Deposit date: | 2024-07-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.046594 Å) | | Cite: | Staphylococcus aureus MazF in complex with nanobody 4.
To Be Published
|
|
8WMN
 
 | | Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (symmetric 20-nt complementary) | | Descriptor: | DNA (62-MER), MAGNESIUM ION, PcrIIC1, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8W6U
 
 | | Ferritin from Ureaplasma diversum soaking in Fe2+ solution for 5 min | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin, ... | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-29 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8YKV
 
 | | Cryo-EM structure of succinate receptor SUCR1 bound to compound 31 | | Descriptor: | (2~{S})-2-[[6-[4-(trifluoromethyloxy)phenyl]pyridin-2-yl]carbonylamino]butanedioic acid, Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
8WYF
 
 | | Cryo-EM structure of DSR2-DSAD1-NAD+ (partial) complex | | Descriptor: | Bacillus phage SPbeta DSAD1 protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WMC
 
 | |
8WXU
 
 | |
8WD0
 
 | | Crystal structure of T2R-TTL-Erianin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methoxy-5-[2-(3,4,5-trimethoxyphenyl)ethyl]phenol, CALCIUM ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-09-14 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The cytotoxic natural compound erianin binds to colchicine site of beta-tubulin and overcomes taxane resistance
Bioorg.Chem., 150, 2024
|
|
8WM6
 
 | | The structure of PSI-CAC(L-14)of R.salina at 2.7 angstroms resolution | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
