3A5U
 
 | | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex | | Descriptor: | DNA (31-MER), Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Manjunath, G.P, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2009-08-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex.
To be Published, 2009
|
|
3RJE
 
 | | Ternary complex of DNA Polymerase Beta with a gapped DNA containing 8odG at template position | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*(8OG)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2011-04-15 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binary complex crystal structure of DNA polymerase beta reveals multiple conformations of the templating 8-oxoguanine lesion
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RJG
 
 | | Binary complex of DNA Polymerase Beta with a gapped DNA containing 8odG:dA base-pair at primer Terminus | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*(8OG)P*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2011-04-15 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binary complex crystal structure of DNA polymerase beta reveals multiple conformations of the templating 8-oxoguanine lesion
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8V5O
 
 | | Tetramer core subcomplex (conformation 3) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.99 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5N
 
 | | Tetramer core subcomplex (conformation 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.56 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5M
 
 | | Tetramer core subcomplex (conformation 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (9.22 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7YPO
 
 | | Cryo-EM structure of baculovirus LEF-3 in complex with ssDNA | | Descriptor: | DNA (28-MER), Lef3 | | Authors: | Yin, J, Fu, Y, Rao, G, Li, Z, Cao, S. | | Deposit date: | 2022-08-04 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural transitions during the cooperative assembly of baculovirus single-stranded DNA-binding protein on ssDNA.
Nucleic Acids Res., 50, 2022
|
|
7YPQ
 
 | | Cryo-EM structure of one baculovirus LEF-3 molecule in complex with ssDNA | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Lef3 | | Authors: | Fu, Y, Rao, G, Yin, J, Li, Z, Cao, S. | | Deposit date: | 2022-08-04 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural transitions during the cooperative assembly of baculovirus single-stranded DNA-binding protein on ssDNA.
Nucleic Acids Res., 50, 2022
|
|
6JIQ
 
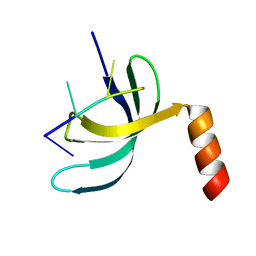 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
6JIP
 
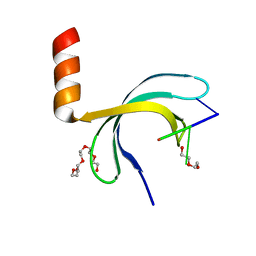 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), PENTAETHYLENE GLYCOL, SP_0782 | | Authors: | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
1VKX
 
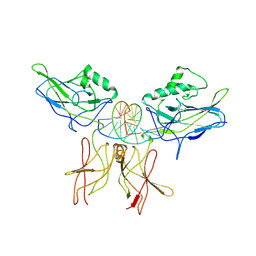 | | CRYSTAL STRUCTURE OF THE NFKB P50/P65 HETERODIMER COMPLEXED TO THE IMMUNOGLOBULIN KB DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*C)-3'), PROTEIN (NF-KAPPA B P50 SUBUNIT), ... | | Authors: | Chen, F, Huang, D.B, Ghosh, G. | | Deposit date: | 1997-09-17 | | Release date: | 1998-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA.
Nature, 391, 1998
|
|
5LTY
 
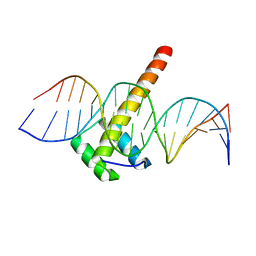 | | Homeobox transcription factor CDX2 bound to methylated DNA | | Descriptor: | DNA (5'-D(P*GP*GP*AP*GP*GP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*CP*CP*TP*CP*C)-3'), Homeobox protein CDX-2 | | Authors: | Morgunova, E, Popov, A, Taipale, J. | | Deposit date: | 2016-09-07 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
2ISP
 
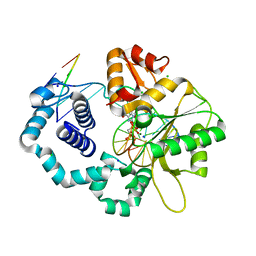 | | Ternary complex of DNA Polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-methylene triphosphate | | Descriptor: | 2'-DEOXY-5'-O-(HYDROXY{[HYDROXY(PHOSPHONOMETHYL)PHOSPHORYL]OXY}PHOSPHORYL)GUANOSINE, 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3', ... | | Authors: | Sucato, C.A, Upton, T.G, Kashemirov, B.A, Martinek, V, Xiang, Y, Beard, W.A. | | Deposit date: | 2006-10-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Modifying the beta,gamma Leaving-Group Bridging Oxygen Alters Nucleotide Incorporation Efficiency, Fidelity, and the Catalytic Mechanism of DNA Polymerase beta.
Biochemistry, 46, 2007
|
|
1ZJM
 
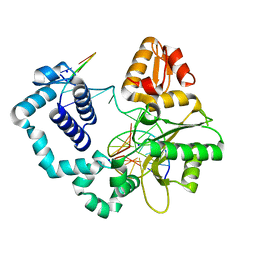 | | Human DNA Polymerase beta complexed with DNA containing an A-A mismatched primer terminus | | Descriptor: | 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3', 5'-D(P*GP*TP*CP*GP*G)-3', DNA polymerase beta, ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2005-04-29 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nucleotide-induced DNA polymerase active site motions accommodating a mutagenic DNA intermediate.
STRUCTURE, 13, 2005
|
|
6GIS
 
 | | Structural basis of human clamp sliding on DNA | | Descriptor: | DNA (5'-D(P*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), Proliferating cell nuclear antigen | | Authors: | De March, M, Merino, N, Barrera-Vilarmau, S, Crehuet, R, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis of human PCNA sliding on DNA.
Nat Commun, 8, 2017
|
|
2C2D
 
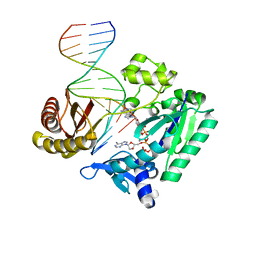 | | Efficient and High Fidelity Incorporation of dCTP Opposite 7,8- Dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA Polymerase Dpo4 | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*C 8OGP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-09-27 | | Release date: | 2005-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Efficient and High Fidelity Incorporation of Dctp Opposite 7,8-Dihydro-8-Oxodeoxyguanosine by Sulfolobus Solfataricus DNA Polymerase Dpo4
J.Biol.Chem., 281, 2006
|
|
5T1J
 
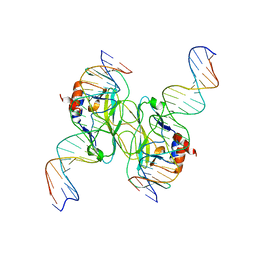 | | Crystal Structure of the Tbox DNA binding domain of the transcription factor T-bet | | Descriptor: | DNA, T-box transcription factor TBX21 | | Authors: | Liu, C.F, Brandt, G.S, Hoang, Q, Hwang, E.S, Naumova, N, Lazarevic, V, Dekker, J, Glimcher, L.H, Ringe, D, Petsko, G.A. | | Deposit date: | 2016-08-19 | | Release date: | 2016-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.947 Å) | | Cite: | Crystal structure of the DNA binding domain of the transcription factor T-bet suggests simultaneous recognition of distant genome sites.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1DU0
 
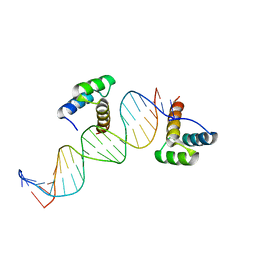 | | ENGRAILED HOMEODOMAIN Q50A VARIANT DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*TP*AP*AP*TP*TP*AP*CP*AP*TP*GP*GP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*TP*AP*CP*CP*TP*AP*A)-3'), ENGRAILED HOMEODOMAIN | | Authors: | Grant, R.A, Rould, M.A, Klemm, J.D, Pabo, C.O. | | Deposit date: | 2000-01-13 | | Release date: | 2000-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the role of glutamine 50 in the homeodomain-DNA interface: crystal structure of engrailed (Gln50 --> ala) complex at 2.0 A.
Biochemistry, 39, 2000
|
|
1XBR
 
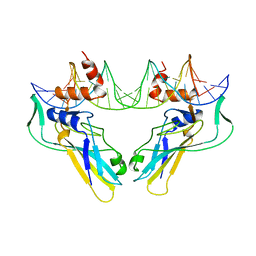 | | T DOMAIN FROM XENOPUS LAEVIS BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*TP*CP*AP*CP*AP*CP*CP*TP*AP*GP*GP*TP*G P*TP*GP*AP*AP*AP* TP*T)-3'), PROTEIN (T PROTEIN) | | Authors: | Muller, C.W. | | Deposit date: | 1997-07-16 | | Release date: | 1998-01-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor.
Nature, 389, 1997
|
|
3U6Y
 
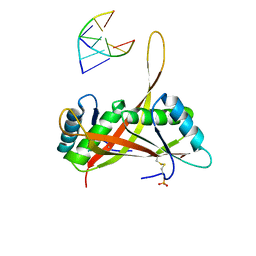 | |
2LKX
 
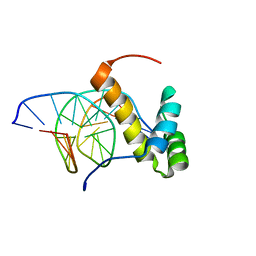 | | NMR structure of the homeodomain of Pitx2 in complex with a TAATCC DNA binding site | | Descriptor: | DNA (5'-D(*CP*GP*GP*GP*GP*AP*TP*TP*AP*GP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*CP*TP*AP*AP*TP*CP*CP*CP*CP*G)-3'), Pituitary homeobox 3 | | Authors: | Baird-Titus, J.M, Doerdelmann, T, Chaney, B.A, Clark-Baldwin, K, Dave, V, Ma, J. | | Deposit date: | 2011-10-21 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the K50 class homeodomain PITX2 bound to DNA and implications for mutations that cause Rieger syndrome
Biochemistry, 44, 2005
|
|
2FMS
 
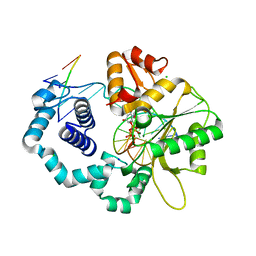 | | DNA Polymerase beta with a gapped DNA substrate and dUMPNPP with magnesium in the catalytic site | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Krahn, J.M, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Magnesium-induced assembly of a complete DNA polymerase catalytic complex.
Structure, 14, 2006
|
|
1L3V
 
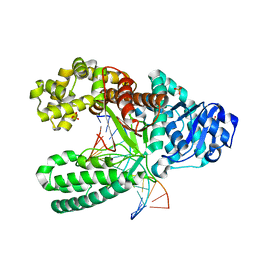 | | Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 15 base pairs of duplex DNA following addition of dTTP, dATP, dCTP, and dGTP residues. | | Descriptor: | 5'-D(*GP*AP*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*AP*CP*GP*TP*C)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2FMP
 
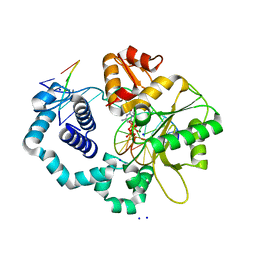 | | DNA Polymerase beta with a terminated gapped DNA substrate and ddCTP with sodium in the catalytic site | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3', ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Krahn, J.M, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Magnesium-induced assembly of a complete DNA polymerase catalytic complex.
Structure, 14, 2006
|
|
1ZJN
 
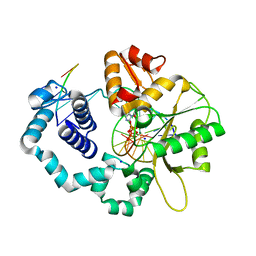 | | Human DNA Polymerase beta complexed with DNA containing an A-A mismatched primer terminus with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2005-04-29 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Nucleotide-induced DNA polymerase active site motions accommodating a mutagenic DNA intermediate.
STRUCTURE, 13, 2005
|
|
