8B4B
 
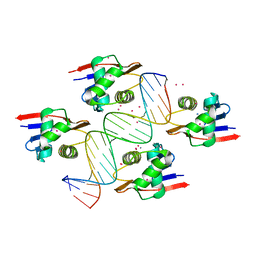 | | ToxR bacterial transcriptional regulator bound to 19 bp ompU promoter DNA | | Descriptor: | AMMONIUM ION, CADMIUM ION, Cholera toxin transcriptional activator, ... | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7UY3
 
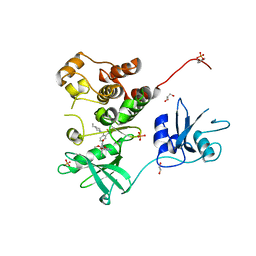 | | Crystal structure of human Fgr tyrosine kinase in complex with TL02-59 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(6,7-dimethoxyquinazolin-4-yl)oxy]-N-{4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-4-methylbenzamide, GLYCEROL, ... | | Authors: | Du, S, Alvarado, J.J, Smithgall, T.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | ATP-site inhibitors induce unique conformations of the acute myeloid leukemia-associated Src-family kinase, Fgr.
Structure, 30, 2022
|
|
6PAZ
 
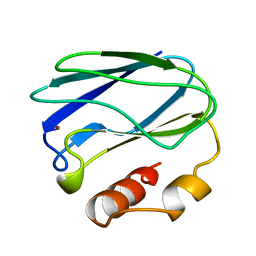 | | OXIDIZED MUTANT P80I PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-21 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
6ZRY
 
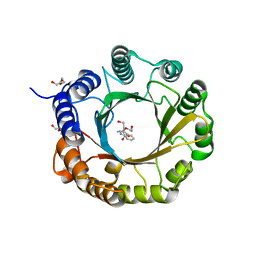 | | 6-dimethylallyl tryptophan synthase | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, DMATS type aromatic prenyltransferase, ... | | Authors: | Ostertag, E, Stehle, T, Zocher, G. | | Deposit date: | 2020-07-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Reprogramming Substrate and Catalytic Promiscuity of Tryptophan Prenyltransferases.
J.Mol.Biol., 433, 2020
|
|
8B4D
 
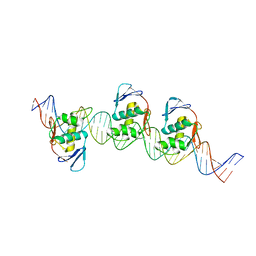 | | ToxR bacterial transcriptional regulator bound to 40 bp toxT promoter DNA | | Descriptor: | Cholera toxin transcriptional activator, DNA (40-MER) | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6PCL
 
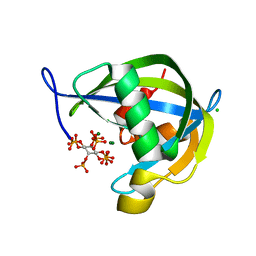 | | Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 1 in complex with 5-IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Dollins, D.E, Neubauer, J, Dong, J, York, J.D. | | Deposit date: | 2019-06-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7UY0
 
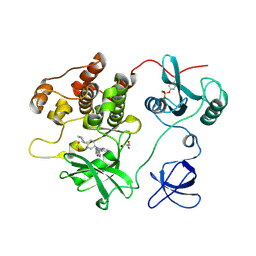 | | Crystal structure of human Fgr tyrosine kinase in complex with A-419259 | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CHLORIDE ION, GLYCEROL, ... | | Authors: | Du, S, Alvarado, J.J, Smithgall, T.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | ATP-site inhibitors induce unique conformations of the acute myeloid leukemia-associated Src-family kinase, Fgr.
Structure, 30, 2022
|
|
8PFC
 
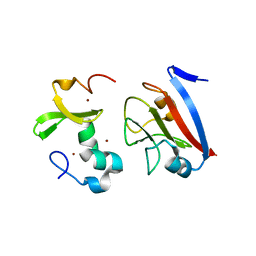 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the zinc finger domain of SPL5 from Arabidopsis thaliana | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 5, ZINC ION | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6ZNN
 
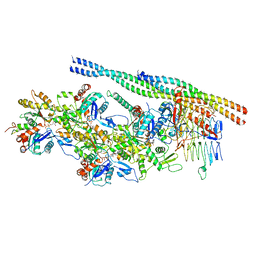 | | The pointed end complex of dynactin bound to Hook3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Lau, C.K, Lacey, S.E, Carter, A.P. | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM reveals the complex architecture of dynactin's shoulder region and pointed end.
Embo J., 40, 2021
|
|
7UIH
 
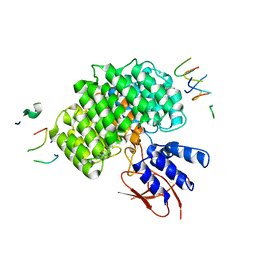 | | PSMD2 Structure | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 2, Fab 14 HC CDRs, Fab 14 LC CDRs, ... | | Authors: | Johnson, M.C, Bashore, C, Ciferri, C, Dueber, E.C. | | Deposit date: | 2022-03-29 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Targeted degradation via direct 26S proteasome recruitment.
Nat.Chem.Biol., 19, 2023
|
|
6P5X
 
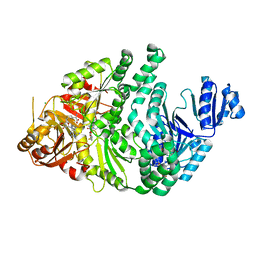 | | Sirohydrochlorin-bound S. typhimurium siroheme synthase | | Descriptor: | 3,3',3'',3'''-[(7S,8S,12S,13S)-3,8,13,17-tetrakis(carboxymethyl)-8,13-dimethyl-7,8,12,13-tetrahydroporphyrin-2,7,12,18-tetrayl]tetrapropanoic acid, CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Pennington, J.M, Stroupe, M.E. | | Deposit date: | 2019-05-31 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Siroheme synthase orients substrates for dehydrogenase and chelatase activities in a common active site.
Nat Commun, 11, 2020
|
|
8V5M
 
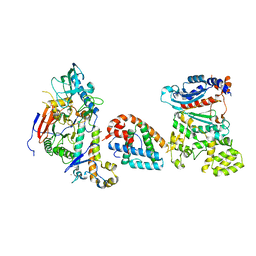 | | Tetramer core subcomplex (conformation 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (9.22 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5LN3
 
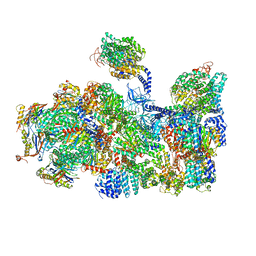 | | The human 26S Proteasome at 6.8 Ang. | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Schweitzer, A, Beck, F, Sakata, E, Unverdorben, P. | | Deposit date: | 2016-08-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular Details Underlying Dynamic Structures and Regulation of the Human 26S Proteasome.
Mol. Cell Proteomics, 16, 2017
|
|
8V5O
 
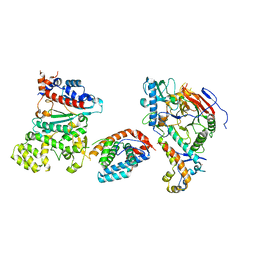 | | Tetramer core subcomplex (conformation 3) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.99 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HZM
 
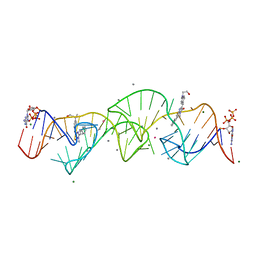 | | A new fluorescent RNA aptamer bound with N, manganese soak | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8V5N
 
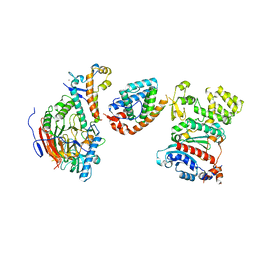 | | Tetramer core subcomplex (conformation 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.56 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6ZWC
 
 | | Z-SBTub2 photoswitch bound to tubulin-DARPin D1 complex | | Descriptor: | 2-[2-(3,4,5-trimethoxyphenyl)ethyl]-1,3-benzothiazole, Designed Ankyrin Repeat Protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wranik, M, Weinert, T, Olieric, N, Gao, L, Kraus, Y.C.M, Bingham, R, Ntouliou, E, Ahlfeld, J, Thorn-Seshold, O, Steinmetz, M.O, Standfuss, J. | | Deposit date: | 2020-07-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A Robust, GFP-Orthogonal Photoswitchable Inhibitor Scaffold Extends Optical Control over the Microtubule Cytoskeleton.
Cell Chem Biol, 28, 2021
|
|
6ZOL
 
 | | SARS-CoV-2-Nsp1-40S complex, focused on head | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S12, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M.L, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8HZE
 
 | | A new fluorescent RNA aptamer bound with N | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8B2L
 
 | | Cryo-EM structure of the plant 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 30S ribosomal protein S15, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-09-14 | | Release date: | 2023-08-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8HZD
 
 | | A new fluorescent RNA aptamer bound with N618 | | Descriptor: | 4-[(~{E})-2-[(4~{Z})-4-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]methylidene]-1-methyl-5-oxidanylidene-imidazol-2-yl]ethenyl]benzenecarbonitrile, MAGNESIUM ION, RNA (36-MER) | | Authors: | Song, Q.Q, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
6OMF
 
 | | CryoEM structure of SigmaS-transcription initiation complex with activator Crl | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Jaramillo Cartagena, A, Darst, S.A, Campbell, E.A. | | Deposit date: | 2019-04-18 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for transcription activation by Crl through tethering of sigmaSand RNA polymerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6XD3
 
 | | Structure of the human CAK in complex with THZ1 | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Greber, B.J, Perez-Bertoldi, J.M, Lim, K, Iavarone, A.T, Toso, D.B, Nogales, E. | | Deposit date: | 2020-06-09 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The cryoelectron microscopy structure of the human CDK-activating kinase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8HZF
 
 | | A new fluorescent RNA aptamer bound with N565 | | Descriptor: | (5~{Z})-5-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]methylidene]-2-[(~{E})-2-(4-hydroxyphenyl)ethenyl]-3-methyl-imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
6AN8
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 8.0 | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA PRIMER (5'- D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*GP)-3'), DNA TEMPLATE (5'- D(*AP*TP*GP*AP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-08-12 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
