6ZD6
 
 | | Structure of apo telomerase from Candida Tropicalis | | Descriptor: | Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-13 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
3F6P
 
 | | Crystal Structure of unphosphorelated receiver domain of YycF | | Descriptor: | Transcriptional regulatory protein yycF | | Authors: | Zhao, H, Tang, L. | | Deposit date: | 2008-11-06 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Preliminary crystallographic studies of the regulatory domain of response regulator YycF from an essential two-component signal transduction system.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
6ZDP
 
 | | Structure of telomerase from Candida Tropicalis in complexe with TWJ fragment of telomeric RNA | | Descriptor: | Chains: B, POTASSIUM ION, Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-15 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
1Z5X
 
 | | hemipteran ecdysone receptor ligand-binding domain complexed with ponasterone A | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor ligand binding domain, PHOSPHATE ION, ... | | Authors: | Carmichael, J.A, Lawrence, M.C, Graham, L.D, Pilling, P.A, Epa, V.C, Noyce, L, Lovrecz, G, Winkler, D.A, Pawlak-Skrzecz, A. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | The X-ray structure of a hemipteran ecdysone receptor ligand-binding domain: comparison with a lepidopteran ecdysone receptor ligand-binding domain and implications for insecticide design.
J.Biol.Chem., 280, 2005
|
|
3U9Q
 
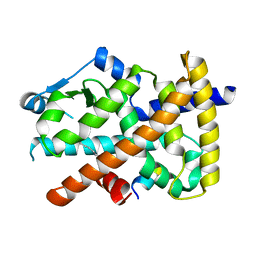 | |
3TKC
 
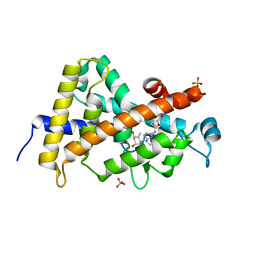 | | Design, Synthesis, Evaluation and Structure of Vitamin D Analogues with Furan Side Chains | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha,20S)-20-[5-(1-hydroxy-1-methylethyl)furan-2-yl]-9,10-secopregna-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Huet, T, Moras, D, Rochel, N. | | Deposit date: | 2011-08-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis, evaluation, and structure of vitamin D analogues with furan side chains.
Chemistry, 18, 2012
|
|
1Z0X
 
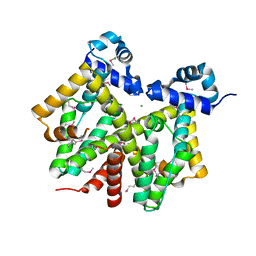 | | Crystal structure of transcriptional regulator, tetR Family from Enterococcus faecalis V583 | | Descriptor: | CHLORIDE ION, transcriptional regulator, TetR family | | Authors: | Chang, C, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-02 | | Release date: | 2005-04-19 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of transcriptional regulator, tetR Family from Enterococcus faecalis V583
To be Published
|
|
8FH3
 
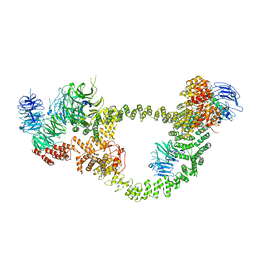 | | Human IFT-A complex structures provide molecular insights into ciliary transport | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Tubby-related protein 3, ... | | Authors: | Jiang, M, Palicharla, V.R, Miller, D, Hwang, S.H, Zhu, H, Hixson, P, Mukhopadhyay, S, Sun, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Human IFT-A complex structures provide molecular insights into ciliary transport.
Cell Res., 33, 2023
|
|
8Q3O
 
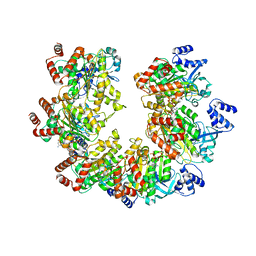 | |
8Q3Q
 
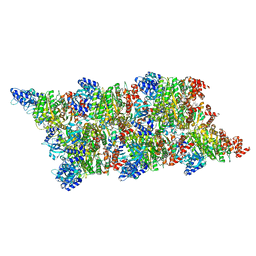 | |
8Q3N
 
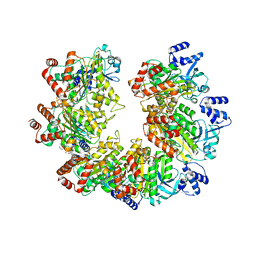 | |
8Q3P
 
 | |
5WA9
 
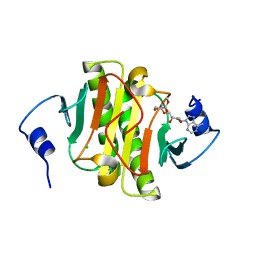 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside D-Ala phosphoramidate substrate complex | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[(2~{R})-1-methoxy-1-oxidanylidene-propan-2-yl]phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2017-06-26 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
8PJ7
 
 | | MLLT3 in complex with compound PFI-6 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Raux, B, Diaz-Saez, L, Huber, K.V.M, Fedorov, O, Owen, D.R, Londregan, A.T, Bountra, C, Edwards, A, Arrowsmith, C. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery of PFI-6, a small-molecule chemical probe for the YEATS domain of MLLT1 and MLLT3.
Bioorg.Med.Chem.Lett., 98, 2023
|
|
6Z2Z
 
 | |
6Z2U
 
 | |
1ZEO
 
 | | Crystal Structure of Human PPAR-gamma Ligand Binding Domain Complexed with an Alpha-Aryloxyphenylacetic Acid Agonist | | Descriptor: | (2S)-(4-ISOPROPYLPHENYL)[(2-METHYL-3-OXO-5,7-DIPROPYL-2,3-DIHYDRO-1,2-BENZISOXAZOL-6-YL)OXY]ACETATE, Peroxisome proliferator activated receptor gamma | | Authors: | Shi, G.Q, Dropinski, J.F, McKeever, B.M, Adams, A.D, MacNaul, K.L, Elbrecht, A, Berger, J.P, Zhou, G, Doebber, T.W. | | Deposit date: | 2005-04-19 | | Release date: | 2006-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and Synthesis of alpha-Aryloxyphenylacetic Acid Derivatives: A Novel Class of PPAR alpha/gamma Dual Agonists with Potent Antihyperglycemic and Lipid Modulating Activity
J.Med.Chem., 48, 2005
|
|
3TW2
 
 | | High resolution structure of human histidine triad nucleotide-binding protein 1 (hHINT1)/AMP complex in a monoclinic space group | | Descriptor: | ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Dolot, R.M, Wlodarczyk, A, Ozga, M, Krakowiak, A, Nawrot, B. | | Deposit date: | 2011-09-21 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A new crystal form of human histidine triad nucleotide-binding protein 1 (hHINT1) in complex with adenosine 5'-monophosphate at 1.38 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6WM8
 
 | |
6WQ1
 
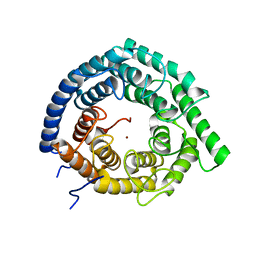 | | Eukaryotic LanCL2 protein | | Descriptor: | LanC-like protein 2, ZINC ION | | Authors: | Nair, S.K, Garg, N. | | Deposit date: | 2020-04-28 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | LanCLs add glutathione to dehydroamino acids generated at phosphorylated sites in the proteome.
Cell, 184, 2021
|
|
3NRW
 
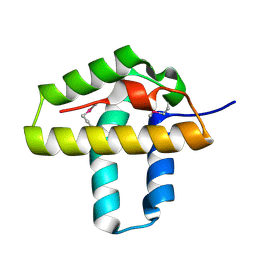 | | Crystal Structure of the N-terminal domain of Phage integrase/site-specific recombinase (tnp) from Haloarcula marismortui, Northeast Structural Genomics Consortium Target HmR208A | | Descriptor: | Phage integrase/site-specific recombinase | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Northeast Structural Genomics Consortium Target HmR208A
To be Published
|
|
6WC6
 
 | |
7AAP
 
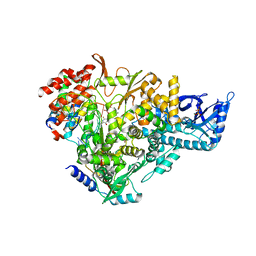 | | Nsp7-Nsp8-Nsp12 SARS-CoV2 RNA-dependent RNA polymerase in complex with template:primer dsRNA and favipiravir-RTP | | Descriptor: | MAGNESIUM ION, Non-structural protein 12, Non-structural protein 7, ... | | Authors: | Naydenova, K, Muir, K.W, Wu, L.F, Zhang, Z, Coscia, F, Peet, M, Castro-Hartman, P, Qian, P, Sader, K, Dent, K, Kimanius, D, Sutherland, J.D, Lowe, J, Barford, D, Russo, C.J. | | Deposit date: | 2020-09-04 | | Release date: | 2020-09-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7THM
 
 | | SARS-CoV-2 nsp12/7/8 complex with a native N-terminus nsp9 | | Descriptor: | MANGANESE (II) ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Osinski, A, Tagliabracci, V.S, Chen, Z, Li, Y. | | Deposit date: | 2022-01-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The mechanism of RNA capping by SARS-CoV-2.
Nature, 609, 2022
|
|
6ODB
 
 | | Crystal structure of HDAC8 in complex with compound 3 | | Descriptor: | GLYCEROL, Histone deacetylase 8, N-{2-[(1E)-3-(hydroxyamino)-3-oxoprop-1-en-1-yl]phenyl}-2-phenoxybenzamide, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
