4H49
 
 | | Crystal structure of the catalytic domain of MMP-12 in complex with a twin inhibitor. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Antoni, C, Stura, E.A, Vera, L, Nuti, E, Carafa, L, Cassar-Lajeunesse, E, Dive, V, Rossello, A. | | Deposit date: | 2012-09-17 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
4GTD
 
 | | T. Maritima FDTS (E144R mutant) with FAD and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Lesley, S.A, Kohen, A, Prabhakar, A. | | Deposit date: | 2012-08-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Folate binding site of flavin-dependent thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MBT
 
 | | Structure of monomeric Blc from E. coli | | Descriptor: | Outer membrane lipoprotein blc | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2010-03-26 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analyses reveal a monomeric state of the bacterial lipocalin Blc.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4GV9
 
 | | Lassa nucleoprotein C-terminal domain in complex with triphosphated dsRNA soaking for 5 min | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, RNA (5'-R(*(GTP)P*GP*GP*C)-3'), ... | | Authors: | Jiang, X, Huang, Q, Wang, W, Dong, H, Ly, H, Liang, Y, Dong, C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structures of Arenaviral Nucleoproteins with Triphosphate dsRNA Reveal a Unique Mechanism of Immune Suppression.
J.Biol.Chem., 288, 2013
|
|
6TTT
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 2 (ASI_M3M_140) | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-~{N}-methyl-3,4-bis(oxidanyl)oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
1K3J
 
 | | Refined NMR Structure of the FHA1 Domain of Yeast Rad53 | | Descriptor: | Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1K58
 
 | | Crystal Structure of Human Angiogenin Variant D116H | | Descriptor: | Angiogenin | | Authors: | Leonidas, D.D, Shapiro, R, Subbarao, G.V, Russo, A, Acharya, K.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies on the role of the C-terminal segment of human angiogenin in defining enzymatic potency.
Biochemistry, 41, 2002
|
|
4HB4
 
 | |
1K5H
 
 | | 1-deoxy-D-xylulose-5-phosphate reductoisomerase | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate reductoisomerase | | Authors: | Reuter, K, Sanderbrand, S, Jomaa, H, Wiesner, J, Steinbrecher, I, Beck, E, Hintz, M, Klebe, G, Stubbs, M.T. | | Deposit date: | 2001-10-10 | | Release date: | 2002-02-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 1-deoxy-D-xylulose-5-phosphate reductoisomerase, a crucial enzyme in the non-mevalonate pathway of isoprenoid biosynthesis.
J.Biol.Chem., 277, 2002
|
|
6TLU
 
 | | HUMAN CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 4,5-DIBROMOBENZOTRIAZOLE | | Descriptor: | 6,7-bis(bromanyl)-1~{H}-benzotriazole, Casein kinase II subunit alpha, SODIUM ION | | Authors: | Czapinska, H, Piasecka, A, Winiewska-Szajewska, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2.
J.Phys.Chem.B, 125, 2021
|
|
6TM7
 
 | | Human 14-3-3 sigma isoform in complex with PLP | | Descriptor: | 14-3-3 protein sigma, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-12-03 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of Phosphate-Containing Compounds as New Inhibitors of 14-3-3/c-Abl Protein-Protein Interaction.
Acs Chem.Biol., 15, 2020
|
|
4H1Q
 
 | | Crystal structure of mutant MMP-9 catalytic domain in complex with a twin inhibitor. | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-9, N-(4-{[(3R)-3-[(biphenyl-4-ylsulfonyl)(propan-2-yloxy)amino]-4-(hydroxyamino)-4-oxobutyl]amino}-4-oxobutyl)-N'-(4-{[(3S)-3-[(biphenyl-4-ylsulfonyl)(propan-2-yloxy)amino]-4-(hydroxyamino)-4-oxobutyl]amino}-4-oxobutyl)benzene-1,3-dicarboxamide, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Catalani, M.P, Dive, V, Rossello, A. | | Deposit date: | 2012-09-11 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
3M6M
 
 | | Crystal structure of RpfF complexed with REC domain of RpfC | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Cheng, Z, Lim, S.C, Qamra, R, Song, H. | | Deposit date: | 2010-03-16 | | Release date: | 2010-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of the Sensor-Synthase Interaction in Autoinduction of the Quorum Sensing Signal DSF Biosynthesis
Structure, 18, 2010
|
|
8GIX
 
 | | Chaetomium thermophilum Hir3 | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Histone transcription regulator 3 homolog | | Authors: | Szurgot, M.R, van Eeuwen, T, Kim, H.J, Marmorstein, R. | | Deposit date: | 2023-03-14 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 84, 2024
|
|
1K59
 
 | | Crystal Structure of Human Angiogenin Variant Q117G | | Descriptor: | CITRIC ACID, angiogenin | | Authors: | Leonidas, D.D, Shapiro, R, Subbarao, G.V, Russo, A, Acharya, K.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies on the role of the C-terminal segment of human angiogenin in defining enzymatic potency.
Biochemistry, 41, 2002
|
|
1K7K
 
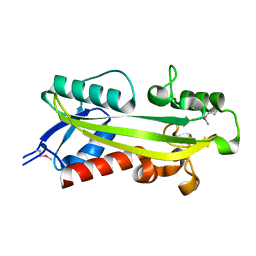 | | crystal structure of RdgB- inosine triphosphate pyrophosphatase from E. coli | | Descriptor: | Hypothetical protein yggV | | Authors: | Sanishvili, R, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-19 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
4H7O
 
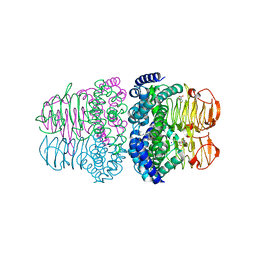 | |
1K3F
 
 | | Uridine Phosphorylase from E. coli, Refined in the Monoclinic Crystal Lattice | | Descriptor: | uridine phosphorylase | | Authors: | Morgunova, E.Yu, Mikhailov, A.M, Popov, A.N, Blagova, E.V, Smirnova, E.A, Vainshtein, B.K, Mao, C, Armstrong, S.R, Ealick, S.E, Komissarov, A.A, Linkova, E.V, Burlakova, A.A, Mironov, A.S, Debabov, V.G. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic structure at 2.5 A resolution of uridine phosphorylase from E. coli as refined in the monoclinic crystal lattice.
FEBS Lett., 367, 1995
|
|
1K9S
 
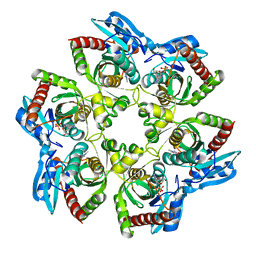 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM E. COLI IN COMPLEX WITH FORMYCIN A DERIVATIVE AND PHOSPHATE | | Descriptor: | 2-(7-AMINO-6-METHYL-3H-PYRAZOLO[4,3-D]PYRIMIDIN-3-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 2-HYDROXYMETHYL-5-(7-METHYLAMINO-3H-PYRAZOLO[4,3-D]PYRIMIDIN-3-YL)-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, ... | | Authors: | Koellner, G, Bzowska, A, Wielgus-Kutrowska, B, Luic, M, Steiner, T, Saenger, W, Stepinski, J. | | Deposit date: | 2001-10-30 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Open and closed conformation of the E. coli purine nucleoside phosphorylase active center and implications for the catalytic mechanism.
J.Mol.Biol., 315, 2002
|
|
1K44
 
 | |
6TQZ
 
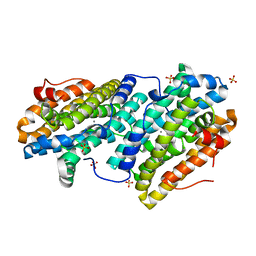 | |
4HAZ
 
 | | Crystal structure of CRM1 inhibitor Leptomycin B in complex with CRM1(R543S,K548E,K579Q)-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1K5B
 
 | | Crystal Structure of Human Angiogenin Variant des(121-123) | | Descriptor: | Angiogenin, CITRIC ACID | | Authors: | Leonidas, D.D, Shapiro, R, Subbarao, G.V, Russo, A, Acharya, K.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies on the role of the C-terminal segment of human angiogenin in defining enzymatic potency.
Biochemistry, 41, 2002
|
|
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
1K7H
 
 | | CRYSTAL STRUCTURE OF SHRIMP ALKALINE PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALKALINE PHOSPHATASE, MALEIC ACID, ... | | Authors: | De Backer, M.E, Mc Sweeney, S, Rasmussen, H.B, Riise, B.W, Lindley, P, Hough, E. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The 1.9 A Crystal Structure of Heat-Labile Shrimp Alkaline Phosphatase
J.Mol.Biol., 318, 2002
|
|
