8GS6
 
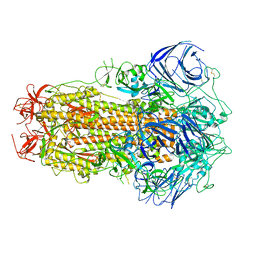 | | Structure of the SARS-CoV-2 BA.2.75 spike glycoprotein (closed state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Anraku, Y, Tabata-Sasaki, K, Kita, S, Fukuhara, H, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2022-09-05 | | Release date: | 2022-10-26 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron BA.2.75 variant.
Cell Host Microbe, 30, 2022
|
|
6W0C
 
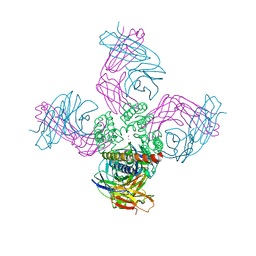 | | Open-gate KcsA soaked in 4 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.556 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6W1V
 
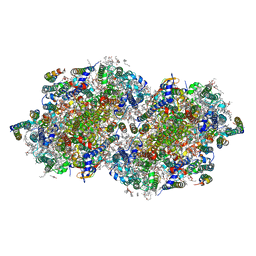 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4DOY
 
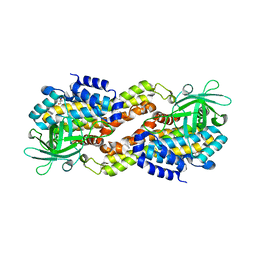 | | Crystal structure of Dibenzothiophene desulfurization enzyme C | | Descriptor: | Dibenzothiophene desulfurization enzyme C, GLYCEROL | | Authors: | Liu, S, Zhang, C, Zhu, D, Gu, L. | | Deposit date: | 2012-02-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Crystal structure of DszC from Rhodococcus sp. XP at 1.79 angstrom
Proteins, 82, 2014
|
|
8GVK
 
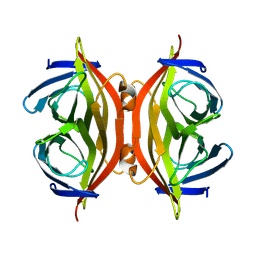 | |
6W3U
 
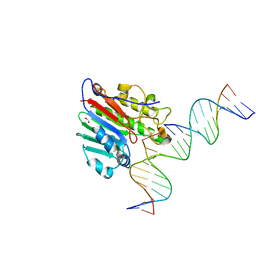 | | APE1 exonuclease substrate complex R237C | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
6W2P
 
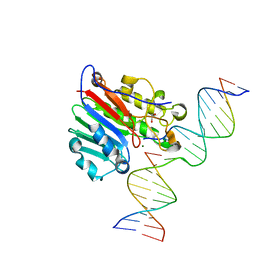 | | APE1 endonuclease product complex L104R | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
5UVI
 
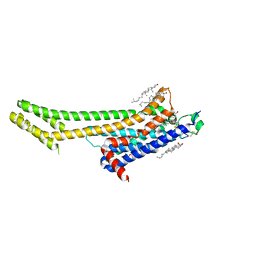 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
IUCrJ, 4, 2017
|
|
5UYL
 
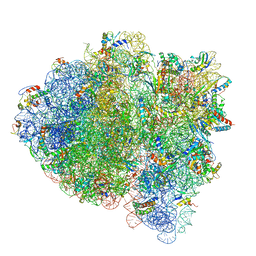 | | 70S ribosome bound with cognate ternary complex base-paired to A site codon (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity.
Nature, 546, 2017
|
|
8GU4
 
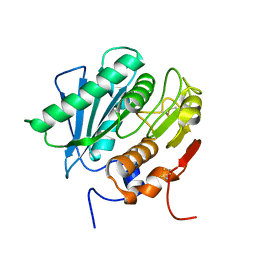 | | Poly(ethylene terephthalate) hydrolase (IsPETase)-linker | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Xiao, Y.J, Wang, Z.F. | | Deposit date: | 2022-09-09 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biodegradation of highly crystallized poly(ethylene terephthalate) through cell surface codisplay of bacterial PETase and hydrophobin.
Nat Commun, 13, 2022
|
|
8GU5
 
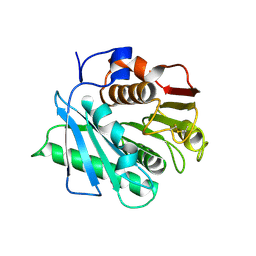 | | Wild type poly(ethylene terephthalate) hydrolase | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Xiao, Y.J, Wang, Z.F. | | Deposit date: | 2022-09-09 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biodegradation of highly crystallized poly(ethylene terephthalate) through cell surface codisplay of bacterial PETase and hydrophobin.
Nat Commun, 13, 2022
|
|
6RQE
 
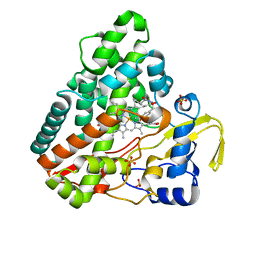 | | CYP121 in complex with 3-acetylene dicyclotyrosine | | Descriptor: | 3-acetylene dicyclotyrosine, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure-Activity Relationships ofcyclo(l-Tyrosyl-l-tyrosine) Derivatives Binding toMycobacterium tuberculosisCYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQO
 
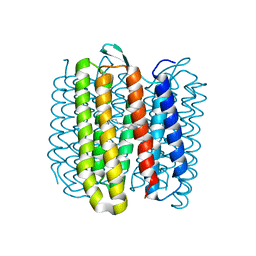 | | Steady-state-SMX activated state structure of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, A, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-16 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
2MUM
 
 | | Solution structure of the PHD domain of Yeast YNG2 | | Descriptor: | Chromatin modification-related protein YNG2, ZINC ION | | Authors: | Taeb, S, Kaustov, L, Lemak, A, Farhadi, S, Sheng, Y. | | Deposit date: | 2014-09-12 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PHD domain of Yeast YNG2
To be Published
|
|
6W7V
 
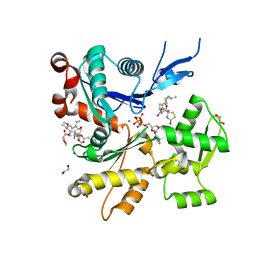 | | Structure of rabbit actin in complex with truncated analog of Mycalolide B | | Descriptor: | (1E,3R,4R,5S,6R,9S,10S,12S)-12-[(4-aminobutanoyl)oxy]-1-[ethyl(formyl)amino]-4,10-dimethoxy-3,5,9,13-tetramethyltetradec-1-en-6-yl (2R)-oxolane-2-carboxylate, 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Allingham, J.S, Deng, X, Trofimova, D. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Truncated Actin-Targeting Macrolide Derivative Blocks Cancer Cell Motility and Invasion of Extracellular Matrix.
J.Am.Chem.Soc., 143, 2021
|
|
4DUM
 
 | | Co-crystal structure of eIF4E with inhibitor | | Descriptor: | (4-{7-[2-(4-chlorophenoxy)ethyl]-2-(methylamino)-6-oxo-6,7-dihydro-1H-purin-8-yl}phenyl)phosphonic acid, 1,2-ETHANEDIOL, Eukaryotic translation initiation factor 4E | | Authors: | Min, X, Johnstone, S, Walker, N, Wang, Z. | | Deposit date: | 2012-02-22 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-Guided Design, Synthesis, and Evaluation of Guanine-Derived Inhibitors of the eIF4E mRNA-Cap Interaction.
J.Med.Chem., 55, 2012
|
|
5V3O
 
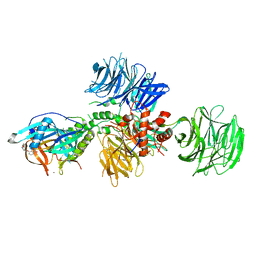 | | Cereblon in complex with DDB1 and CC-220 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Matyskiela, M, Pagarigan, B, Chamberlain, P. | | Deposit date: | 2017-03-07 | | Release date: | 2017-05-03 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Cereblon Modulator (CC-220) with Improved Degradation of Ikaros and Aiolos.
J. Med. Chem., 61, 2018
|
|
2MWE
 
 | | NMR structure of FBP28 WW2 mutant Y438R, L453A DNDC | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Macias, M.J, Scheraga, H, Sunol, D, Todorovski, T. | | Deposit date: | 2014-11-04 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding kinetics of WW domains with the united residue force field for bridging microscopic motions and experimental measurements.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6RMK
 
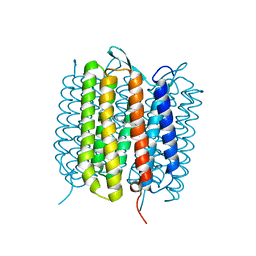 | | Bacteriorhodopsin, dark state, cell 2, refined using the same protocol as sub-ps time delays | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2019-05-07 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
5V56
 
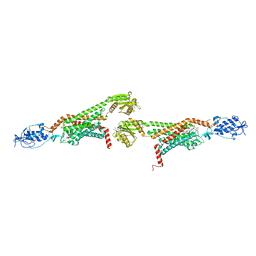 | | 2.9A XFEL structure of the multi-domain human smoothened receptor (with E194M mutation) in complex with TC114 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN MONONUCLEOTIDE, N-methyl-N-[1-[4-(2-methylpyrazol-3-yl)phthalazin-1-yl]piperidin-4-yl]-4-nitro-2-(trifluoromethyl)benzamide, ... | | Authors: | Zhang, X, Zhao, F, Wu, Y, Yang, J, Han, G.W, Zhao, S, Ishchenko, A, Ye, L, Lin, X, Ding, K, Dharmarajan, V, Griffin, P.R, Gati, C, Nelson, G, Hunter, M.S, Hanson, M.A, Cherezov, V, Stevens, R.C, Tan, W, Tao, H, Xu, F. | | Deposit date: | 2017-03-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand.
Nat Commun, 8, 2017
|
|
6RQ8
 
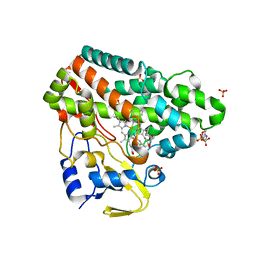 | | CYP121 in complex with 3-iodo dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(4-hydroxyphenyl)methyl]-6-[(3-iodanyl-4-oxidanyl-phenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
4DJK
 
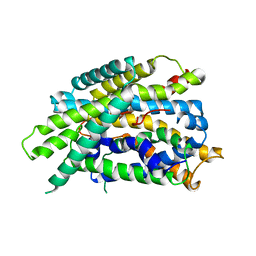 | | Structure of glutamate-GABA antiporter GadC | | Descriptor: | Probable glutamate/gamma-aminobutyrate antiporter | | Authors: | Ma, D, Lu, P.L, Yan, C.Y, Fan, C, Yin, P, Wang, J.W, Shi, Y.G. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structure and mechanism of a glutamate-GABA antiporter
Nature, 483, 2012
|
|
8GT6
 
 | | human STING With agonist HB3089 | | Descriptor: | 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methyl-1H-furo[3,2-e]benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
6RQX
 
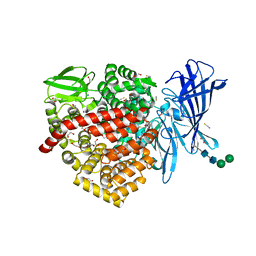 | |
5V4W
 
 | | Human glucokinase in complex with novel indazole activator. | | Descriptor: | (2S)-2-[4-(cyclopropylsulfonyl)-1H-indazol-1-yl]-N-(5-fluoro-1,3-thiazol-2-yl)-3-(oxan-4-yl)propanamide, Glucokinase, IODIDE ION, ... | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2017-03-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of potent and orally active 1,4-disubstituted indazoles as novel allosteric glucokinase activators.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
