8VLX
 
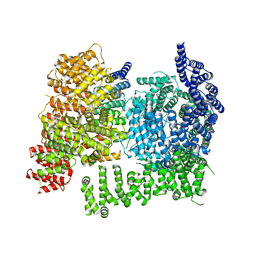 | | HTT in complex with HAP40 and a small molecule. | | Descriptor: | 40-kDa huntingtin-associated protein, Huntingtin, N-[(1S,2R)-2-benzylcyclopentyl]-N'-{1-[(1S)-1-(pyridin-4-yl)ethyl]piperidin-4-yl}urea | | Authors: | Poweleit, N, Boudet, J, Doherty, E. | | Deposit date: | 2024-01-12 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Discovery of a Small Molecule Ligand to the Huntingtin/HAP40 complex
To Be Published
|
|
3N00
 
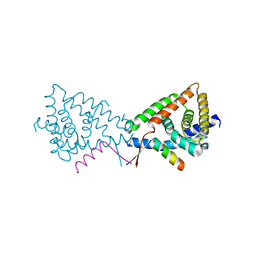 | |
2CW9
 
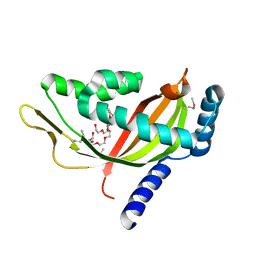 | | Crystal structure of human Tim44 C-terminal domain | | Descriptor: | PENTAETHYLENE GLYCOL, translocase of inner mitochondrial membrane | | Authors: | Handa, N, Kishishita, S, Morita, S, Kinoshita, Y, Nagano, Y, Uda, H, Terada, T, Uchikubo, T, Takemoto, C, Jin, Z, Chrzas, J, Chen, L, Liu, Z.-J, Wang, B.-C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-17 | | Release date: | 2005-12-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human Tim44 C-terminal domain in complex with pentaethylene glycol: ligand-bound form.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
8H40
 
 | | Cryo-EM structure of the transcription activation complex NtcA-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3V
 
 | | Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8BZO
 
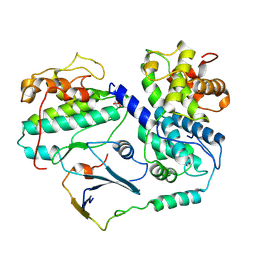 | | Cryo-EM structure of CDK2-CyclinA in complex with p27 from the SCFSKP2 E3 ligase Complex | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E. | | Deposit date: | 2022-12-15 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
3KWY
 
 | |
3TAW
 
 | |
9C5E
 
 | | Covalent Complex Between Parkin Catalytic (Rcat) Domain and Ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase parkin, Polyubiquitin-B, ZINC ION | | Authors: | Connelly, E.M, Rintala-Dempsey, A.C, Gundogdu, M, Freeman, E.A, Koszela, J, Aguirre, J.D, Zhu, G, Kamarainen, O, Tadayon, R, Walden, H, Shaw, G.S. | | Deposit date: | 2024-06-06 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Capturing the catalytic intermediates of parkin ubiquitination.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4ZLF
 
 | | Cellobionic acid phosphorylase - cellobionic acid complex | | Descriptor: | 4-O-beta-D-glucopyranosyl-D-gluconic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
3FN3
 
 | | Dimeric Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Chen, Y, Gao, F, Liu, P, Chu, F, Qi, J, Gao, G.F. | | Deposit date: | 2008-12-23 | | Release date: | 2009-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A dimeric structure of PD-L1: functional units or evolutionary relics?
Protein Cell, 1, 2010
|
|
4ZLI
 
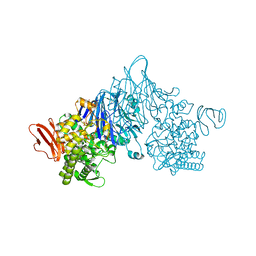 | | Cellobionic acid phosphorylase - 3-O-beta-D-glucopyranosyl-alpha-D-glucopyranuronic acid complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
3G7X
 
 | |
1KD7
 
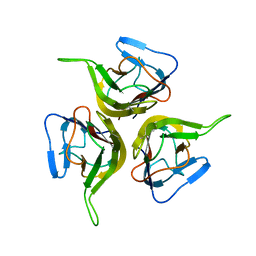 | | Crystal structure of an extracellular domain fragment of human BAFF | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 13B | | Authors: | Karpusas, M, Cachero, T.G, Qian, F, Boriack-Sjodin, A, Mullen, C, Strauch, K, Hsu, Y.-M, Kalled, S.L. | | Deposit date: | 2001-11-12 | | Release date: | 2002-11-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Extracellular Human BAFF, a TNF Family Member that Stimulates B Lymphocytes
J.Mol.Biol., 315, 2002
|
|
2Q02
 
 | |
6NTS
 
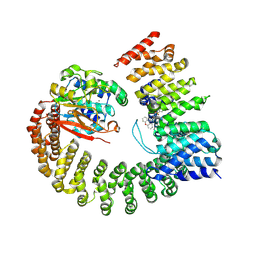 | | Protein Phosphatase 2A (Aalpha-B56alpha-Calpha) holoenzyme in complex with a Small Molecule Activator of PP2A (SMAP) | | Descriptor: | MANGANESE (II) ION, N-[(1R,2R,3S)-2-hydroxy-3-(10H-phenoxazin-10-yl)cyclohexyl]-4-(trifluoromethoxy)benzene-1-sulfonamide, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2019-01-30 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Selective PP2A Enhancement through Biased Heterotrimer Stabilization.
Cell, 181, 2020
|
|
3PCV
 
 | | Crystal structure analysis of human leukotriene C4 synthase at 1.9 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, DODECYL-BETA-D-MALTOSIDE, GLUTATHIONE, ... | | Authors: | Saino, H, Ago, H, Miyano, M. | | Deposit date: | 2010-10-22 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The catalytic architecture of leukotriene C4 synthase with two arginine residues
J.Biol.Chem., 286, 2011
|
|
2EWA
 
 | | Dual binding mode of pyridinylimidazole to MAP kinase p38 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, Mitogen-activated protein kinase 14 | | Authors: | Delarbre, L, Pouzieux, S, Guilloteau, J.-P, Michot, N. | | Deposit date: | 2005-11-02 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NMR characterization of kinase p38 dynamics in free and ligand-bound forms.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
1MQ9
 
 | | Crystal structure of high affinity alphaL I domain with ligand mimetic crystal contact | | Descriptor: | Integrin alpha-L, MANGANESE (II) ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MQA
 
 | | Crystal structure of high affinity alphaL I domain in the absence of ligand or metal | | Descriptor: | Integrin alpha-L | | Authors: | Shimaoka, T, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, Zhang, R, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
2LB2
 
 | | Structure of the second domain of human Nedd4L in complex with a phosphorylated pTPY motif derived from human Smad3 | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like, Mothers against decapentaplegic homolog 3 | | Authors: | Macias, M.J, Aragon, E, Goerner, N, Zaromytidou, A, Xi, Q, Escobedo, A, Massague, J. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A Smad action turnover switch operated by WW domain readers of a phosphoserine code.
Genes Dev., 25, 2011
|
|
2QDR
 
 | |
4BS5
 
 | | MOUSE CATHEPSIN S WITH COVALENT LIGAND | | Descriptor: | (2S,4R)-N-[1-(iminomethyl)cyclopropyl]-4-[2-(trifluoromethyl)phenyl]sulfonyl-pyrrolidine-2-carboxamide, CATHEPSIN S | | Authors: | Banner, D.W, Benz, J, Gsell, B, Stihle, M, Ruf, A, Haap, W. | | Deposit date: | 2013-06-07 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Identification of Potent and Selective Cathepsin S (Cats) Inhibitors Containing Different Central Cyclic Scaffolds.
J.Med.Chem., 56, 2013
|
|
2ZKG
 
 | | Crystal structure of unliganded SRA domain of mouse Np95 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1 | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
4OET
 
 | | Crystal structure of NikZ from Campylobacter jejuni, unliganded form | | Descriptor: | GLYCEROL, Putative peptide ABC-transport system periplasmic peptide-binding protein | | Authors: | Lebrette, H, Cavazza, C. | | Deposit date: | 2014-01-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Promiscuous nickel import in human pathogens: structure, thermodynamics, and evolution of extracytoplasmic nickel-binding proteins.
Structure, 22, 2014
|
|
