5TIL
 
 | |
5TMZ
 
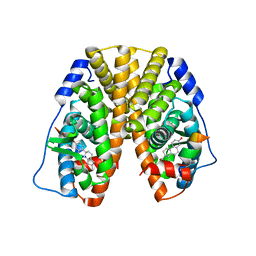 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the estradiol derivative, (8S,9S,13S,14S,17S)-16-(3-methoxybenzyl)-13-methyl-7,8,9,11,12,13,14,15,16,17-decahydro-6H-cyclopenta[a]phenanthrene-3,17-diol | | Descriptor: | (9beta,13alpha,14beta,16alpha,17alpha)-16-[(4-methoxyphenyl)methyl]estra-1,3,5(10)-triene-3,17-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.207 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TN7
 
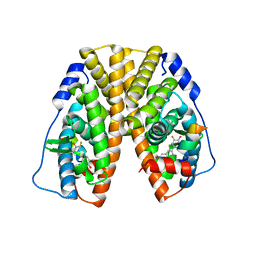 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with (E)-3'-fluoro-4'-hydroxy-3-((hydroxyiminio)methyl)-[1,1'-biphenyl]-4-olate | | Descriptor: | 3-fluoro-3'-[(E)-(hydroxyimino)methyl][1,1'-biphenyl]-4,4'-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Minutolo, F, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
3K7I
 
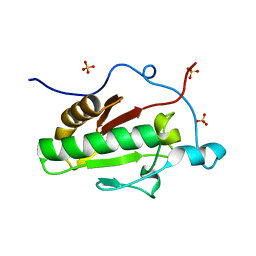 | | Crystal structure of the E131K mutant of the Indian Hedgehog N-terminal signalling domain | | Descriptor: | Indian hedgehog protein, SULFATE ION, ZINC ION | | Authors: | He, Y.-X, Kang, Y, Zhang, W.J, Yu, J, Ma, G, Zhou, C.-Z. | | Deposit date: | 2009-10-13 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.438 Å) | | Cite: | Crystal structure of the E131K mutant of the Indian Hedgehog N-terminal signalling domain
To be Published
|
|
5JOZ
 
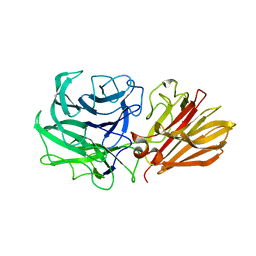 | | Bacteroides ovatus Xyloglucan PUL GH43B | | Descriptor: | CALCIUM ION, Non-reducing end alpha-L-arabinofuranosidase BoGH43B | | Authors: | Hemsworth, G.R, Thompson, A.J, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
5JSN
 
 | | Bcl2-inhibitor complex | | Descriptor: | Apoptosis regulator Bcl-2, Bcl2 inhibitor | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2016-05-09 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computationally designed high specificity inhibitors delineate the roles of BCL2 family proteins in cancer.
Elife, 5, 2016
|
|
5V9P
 
 | | Crystal structure of pyrrolidine amide inhibitor [(3S)-3-(4-bromo-1H-pyrazol-1-yl)pyrrolidin-1-yl][3-(propan-2-yl)-1H-pyrazol-5-yl]methanone (compound 35) in complex with KDM5A | | Descriptor: | Lysine-specific demethylase 5A, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Kiefer, J.R, Liang, J, Vinogradova, M. | | Deposit date: | 2017-03-23 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | From a novel HTS hit to potent, selective, and orally bioavailable KDM5 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5VAM
 
 | | BRAF in Complex with RAF709 | | Descriptor: | N-{2-methyl-5'-(morpholin-4-yl)-6'-[(oxan-4-yl)oxy][3,3'-bipyridin]-5-yl}-3-(trifluoromethyl)benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2017-03-27 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and Discovery of N-(2-Methyl-5'-morpholino-6'-((tetrahydro-2H-pyran-4-yl)oxy)-[3,3'-bipyridin]-5-yl)-3-(trifluoromethyl)benzamide (RAF709): A Potent, Selective, and Efficacious RAF Inhibitor Targeting RAS Mutant Cancers.
J. Med. Chem., 60, 2017
|
|
1R6G
 
 | | Crystal structure of the thyroid hormone receptor beta ligand binding domain in complex with a beta selective compound | | Descriptor: | 2-[3,5-DIBROMO-4-(4-HYDROXY-3-{HYDROXY[(2-PHENYLETHYL)AMINO]METHYL}PHENOXY)PHENYL]ETHANE-1,1-DIOL, Thyroid hormone receptor beta-1 | | Authors: | Hangeland, J.J, Dejneka, T, Friends, T.J, Devasthale, P, Mellstrom, K, Sandberg, J, Grynfarb, M, Doweyko, A.M, Sack, J.S, Einspahr, H, Farnegardh, M, Husman, B, Ljunggren, J, Koehler, K, Sheppard, C, Malm, J, Ryono, D.E. | | Deposit date: | 2003-10-15 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Thyroid receptor ligands. Part 2: Thyromimetics with improved selectivity for the thyroid hormone receptor beta.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1R15
 
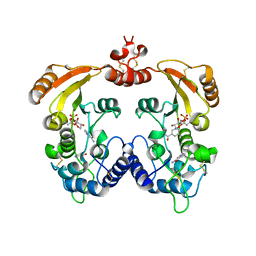 | | Aplysia ADP ribosyl cyclase with bound nicotinamide and R5P | | Descriptor: | ADP-ribosyl cyclase, ANY 5'-MONOPHOSPHATE NUCLEOTIDE, NICOTINAMIDE | | Authors: | Love, M.L, Szebenyi, D.M.E, Kriksunov, I.A, Thiel, D.J, Munshi, C, Graeff, R, Lee, H.C, Hao, Q. | | Deposit date: | 2003-09-23 | | Release date: | 2004-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ADP-ribosyl cyclase; crystal structures reveal a covalent intermediate.
Structure, 12, 2004
|
|
5UIO
 
 | |
5JWD
 
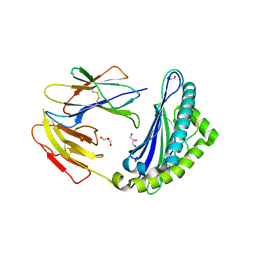 | | Crystal structure of H-2Db in complex with the LCMV-derived GP392-401 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Buratto, J, Badia-Martinez, D, Norstrom, M, Sandalova, T, Achour, A. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of H-2Db in complex with the LCMV-derived peptides GP92 and GP392 explain pleiotropic effects of glycosylation on antigen presentation and immunogenicity.
PLoS ONE, 12, 2017
|
|
1R54
 
 | | Crystal structure of the catalytic domain of human ADAM33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADAM 33, CALCIUM ION, ... | | Authors: | Orth, P, Reicher, P, Wang, W, Prosise, W.W, Yarosh-Tomaine, T, Hammond, G, Xiao, L, Mirza, U.A, Zou, J, Strickland, C, Taremi, S.S. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-12 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structre of the catalytic domain of human ADAM33
J.Mol.Biol., 335, 2004
|
|
1BDA
 
 | |
3K7H
 
 | | Crystal structure of the E95K mutant of the Indian Hedgehog N-terminal signalling domain | | Descriptor: | Indian hedgehog protein, SULFATE ION, ZINC ION | | Authors: | He, Y.-X, Kang, Y, Zhang, W.J, Yu, J, Ma, G, Zhou, C.-Z. | | Deposit date: | 2009-10-13 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the E95K mutant of the Indian Hedgehog N-terminal signalling domain
To be Published
|
|
5V9J
 
 | | Crystal structure of catalytic domain of GLP with MS0105 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Dong, A, Zeng, H, Liu, J, Xiong, Y, Babault, N, Jin, J, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Wu, H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-23 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of catalytic domain of GLP with MS0105
to be published
|
|
5ULK
 
 | |
5VAX
 
 | |
1RFO
 
 | | Trimeric Foldon of the T4 phagehead fibritin | | Descriptor: | whisker antigen control protein | | Authors: | Guthe, S, Kapinos, L, Moglich, A, Meier, S, Kiefhaber, T, Grzesiek, S. | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Very fast folding and association of a trimerization domain from bacteriophage t4 fibritin.
J.Mol.Biol., 337, 2004
|
|
1B3I
 
 | | NMR SOLUTION STRUCTURE OF PLASTOCYANIN FROM THE PHOTOSYNTHETIC PROKARYOTE, PROCHLOROTHRIX HOLLANDICA (MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | COPPER (I) ION, PROTEIN (PLASTOCYANIN) | | Authors: | Babu, C.R, Volkman, B.F, Bullerjahn, G.S. | | Deposit date: | 1998-12-11 | | Release date: | 1999-04-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of plastocyanin from the photosynthetic prokaryote, Prochlorothrix hollandica.
Biochemistry, 38, 1999
|
|
4X3P
 
 | | Sirt2 in complex with a myristoyl peptide | | Descriptor: | 1,2-ETHANEDIOL, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, Y, Zhang, W, Hao, Q. | | Deposit date: | 2014-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deacylation Mechanism by SIRT2 Revealed in the 1'-SH-2'-O-Myristoyl Intermediate Structure.
Cell Chem Biol, 24, 2017
|
|
1AQL
 
 | |
1RU5
 
 | |
1R5A
 
 | | Glutathione S-transferase | | Descriptor: | COPPER (II) ION, GLUTATHIONE SULFONIC ACID, glutathione transferase | | Authors: | Oakley, A.J. | | Deposit date: | 2003-10-09 | | Release date: | 2003-10-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification, characterization and structure of a new Delta class glutathione transferase isoenzyme.
Biochem.J., 388, 2005
|
|
3K05
 
 | |
