6AAH
 
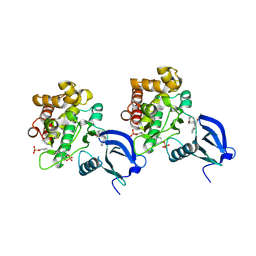 | | Crystal structure of JAK1 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Amano, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
2YFE
 
 | |
4Q0L
 
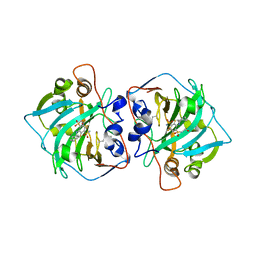 | | Crystal structure of catalytic domain of human carbonic anhydrase isozyme XII with inhibitor | | Descriptor: | 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-02 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
7E5Y
 
 | | Molecular basis for neutralizing antibody 2B11 targeting SARS-CoV-2 RBD | | Descriptor: | 2B11 Fab Heavy chain, 2B11 Fab Light chain, Spike protein S1 | | Authors: | Wu, H, Yu, F, Wang, Q.S, Zhou, H, Wang, W.W, Zhao, T, Pan, Y.B, Yang, X.M. | | Deposit date: | 2021-02-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Screening of potent neutralizing antibodies against SARS-CoV-2 using convalescent patients-derived phage-display libraries.
Cell Discov, 7, 2021
|
|
6LN2
 
 | | Crystal structure of full length human GLP1 receptor in complex with Fab fragment (Fab7F38) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab7F38_heavy chain, Fab7F38_light chain, ... | | Authors: | Wu, F, Yang, L, Hang, K, Laursen, M, Wu, L, Han, G.W, Ren, Q, Roed, N.K, Lin, G, Hanson, M, Jiang, H, Wang, M, Reedtz-Runge, S, Song, G, Stevens, R.C. | | Deposit date: | 2019-12-28 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Full-length human GLP-1 receptor structure without orthosteric ligands.
Nat Commun, 11, 2020
|
|
1WMN
 
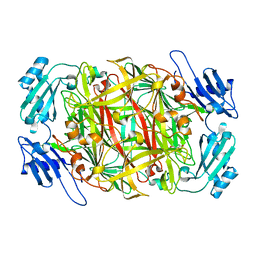 | | Crystal structure of topaquinone-containing amine oxidase activated by cobalt ion | | Descriptor: | COBALT (II) ION, Phenylethylamine oxidase | | Authors: | Okajima, T, Kishishita, S, Chiu, Y.C, Murakawa, T, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | Deposit date: | 2004-07-13 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reinvestigation of metal ion specificity for quinone cofactor biogenesis in bacterial copper amine oxidase
Biochemistry, 44, 2005
|
|
4PZH
 
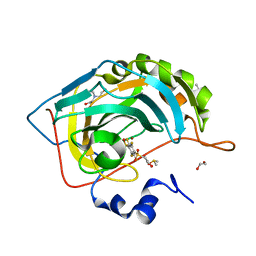 | | Crystal structure of human carbonic anhydrase isozyme II with 2,3,5,6-tetrafluoro-4[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-03-31 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
4Q1W
 
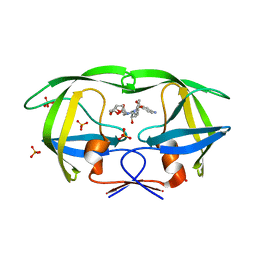 | | Mutations Outside the Active Site of HIV-1 Protease Alter Enzyme Structure and Dynamic Ensemble of the Active Site to Confer Drug Resistance | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, ASPARTYL PROTEASE, ... | | Authors: | Ragland, D.A, Nalam, M.N.L, Cao, H, Nalivaika, E.A, Cai, Y, Kurt-Yilmaz, N, Schiffer, C.A. | | Deposit date: | 2014-04-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Drug resistance conferred by mutations outside the active site through alterations in the dynamic and structural ensemble of HIV-1 protease.
J.Am.Chem.Soc., 136, 2014
|
|
4Q4K
 
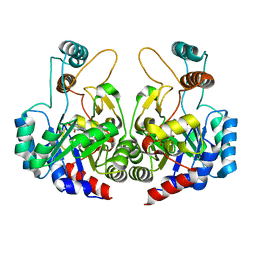 | | Crystal structure of nitronate monooxygenase from Pseudomonas aeruginosa PAO1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitronate Monooxygenase | | Authors: | Salvi, F, Agniswamy, J, Gadda, G, Weber, I.T. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | The Combined Structural and Kinetic Characterization of a Bacterial Nitronate Monooxygenase from Pseudomonas aeruginosa PAO1 Establishes NMO Class I and II.
J.Biol.Chem., 289, 2014
|
|
7JMT
 
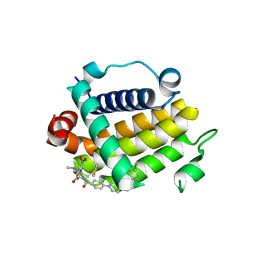 | | Crystal structure of schistosome BCL-2 bound to ABT-737 | | Descriptor: | 4-{4-[(4'-CHLOROBIPHENYL-2-YL)METHYL]PIPERAZIN-1-YL}-N-{[4-({(1R)-3-(DIMETHYLAMINO)-1-[(PHENYLTHIO)METHYL]PROPYL}AMINO)-3-NITROPHENYL]SULFONYL}BENZAMIDE, BCL-2 protein | | Authors: | Smith, N.A, Smith, B.J, Lee, E.F, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2020-08-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimization of Benzothiazole and Thiazole Hydrazones as Inhibitors of Schistosome BCL-2.
Acs Infect Dis., 7, 2021
|
|
3IAW
 
 | |
3IB1
 
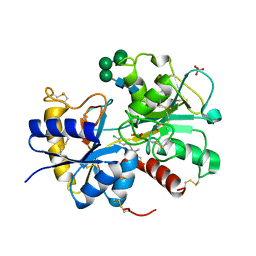 | | Structural basis of the prevention of NSAID-induced damage of the gastrointestinal tract by C-terminal half (C-lobe) of bovine colostrum protein lactoferrin: Binding and structural studies of C-lobe complex with indomethacin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Singh, N, Sinha, M, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis for the prevention of nonsteroidal antiinflammatory drug-induced gastrointestinal tract damage by the C-lobe of bovine colostrum lactoferrin
Biophys.J., 97, 2009
|
|
6LRZ
 
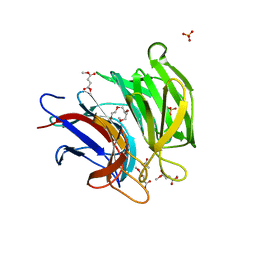 | | Crystal structure of Keap1 in complex with dimethyl fumarate (DMF) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ACETATE ION, Keap1-DC, ... | | Authors: | Padmanabhan, B, Unni, S, Deshmukh, P. | | Deposit date: | 2020-01-16 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the multiple binding modes of Dimethyl Fumarate (DMF) and its analogs to the Kelch domain of Keap1.
Febs J., 288, 2021
|
|
6AAK
 
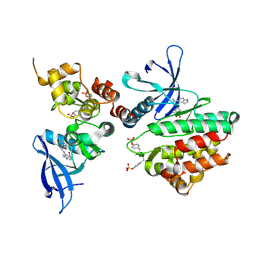 | | Crystal structure of JAK3 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Amano, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
8CDM
 
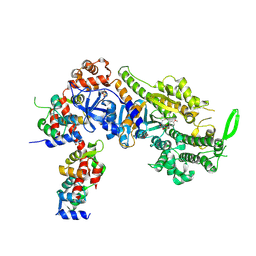 | | Plasmodium falciparum Myosin A full-length, post-rigor state complexed to the inhibitor KNX-002 | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-methoxyphenyl)-~{N}-[(3-thiophen-2-yl-1~{H}-pyrazol-4-yl)methyl]cyclopropan-1-amine, Myosin A tail domain interacting protein, ... | | Authors: | Moussaoui, D, Robblee, J.P, Robert-Paganin, J, Auguin, D, Fisher, F, Fagnant, P.M, MacFarlane, J.E, Mueller-Dieckmann, C, Baum, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2023-01-31 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Plasmodium falciparum Myosin A full-length, post-rigor state complexed to the inhibitor KNX-002
To Be Published
|
|
8I7W
 
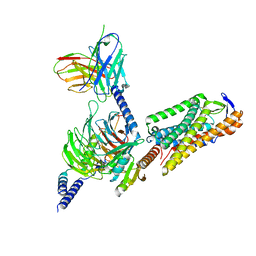 | | Cryo-EM structure of GSK256073 bound human hydroxy-carboxylic acid receptor 2 in complex with Gi heterotrimer | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Park, J.H, Ishimoto, N, Park, S.Y. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for ligand recognition and signaling of hydroxy-carboxylic acid receptor 2.
Nat Commun, 14, 2023
|
|
6ME9
 
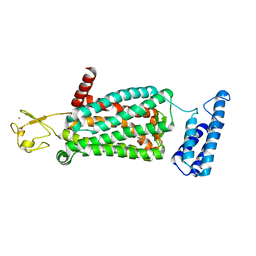 | | XFEL crystal structure of human melatonin receptor MT2 in complex with ramelteon | | Descriptor: | N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}propanamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ZINC ION | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
2JSJ
 
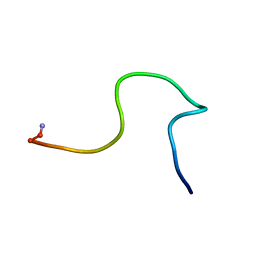 | | Obestatin in water solution | | Descriptor: | Appetite-regulating hormone, Obestatin | | Authors: | D'Ursi, A.M, Scrima, M, Esposito, C, Campiglia, P. | | Deposit date: | 2007-07-05 | | Release date: | 2008-10-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Obestatin conformational features: a strategy to unveil obestatin's biological role?
Biochem.Biophys.Res.Commun., 363, 2007
|
|
3IFO
 
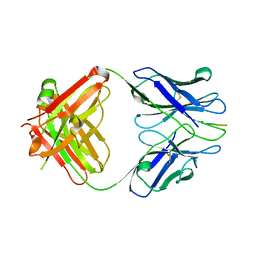 | | X-ray structure of amyloid beta peptide:antibody (Abeta1-7:10D5) complex | | Descriptor: | 10D5 FAB antibody heavy chain, 10D5 FAB antibody light chain, Amyloid beta A4 protein | | Authors: | Weis, W.I, Feinberg, H, Basi, G.S, Schenk, D. | | Deposit date: | 2009-07-24 | | Release date: | 2009-11-17 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural correlates of antibodies associated with acute reversal of amyloid beta-related behavioral deficits in a mouse model of Alzheimer disease.
J.Biol.Chem., 285, 2010
|
|
6MG4
 
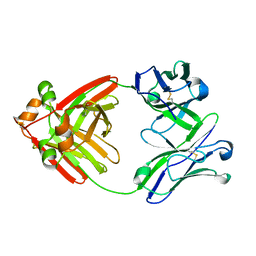 | | Structure of full-length human lambda-6A light chain JTO | | Descriptor: | JTO light chain | | Authors: | Morgan, G.J, Yan, N.L, Mortenson, D.E, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2018-09-12 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Stabilization of amyloidogenic immunoglobulin light chains by small molecules.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2JUT
 
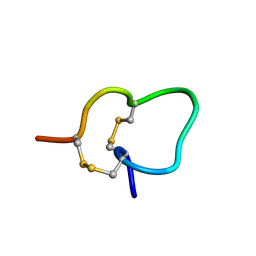 | |
7ENN
 
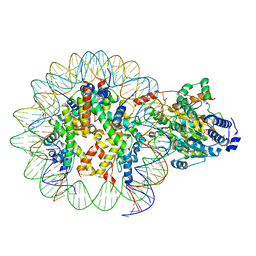 | | The structure of ALC1 bound to the nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromodomain-helicase-DNA-binding protein 1-like, ... | | Authors: | Chen, Z.C, Chen, K.J, Wang, L. | | Deposit date: | 2021-04-18 | | Release date: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of ALC1/CHD1L autoinhibition and the mechanism of activation by the nucleosome.
Nat Commun, 12, 2021
|
|
8IT9
 
 | | Co-crystal structure of FTO bound to 22 | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[(2,6-diethyl-4-pyridin-4-yl-phenyl)amino]-6-(1,4-oxazepan-4-ylmethyl)benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO | | Authors: | Yang, C.-G, Gan, J.H. | | Deposit date: | 2023-03-22 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Rational Design of RNA Demethylase FTO Inhibitors with Enhanced Antileukemia Drug-Like Properties.
J.Med.Chem., 66, 2023
|
|
4LTE
 
 | | Structure of Cysteine-free Human Insulin Degrading Enzyme in Complex with Macrocyclic Inhibitor | | Descriptor: | 2,6-DIAMINO-HEXANOIC ACID AMIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FUMARIC ACID, ... | | Authors: | Foda, Z.H, Seeliger, M.A, Saghatelian, A, Liu, D.R. | | Deposit date: | 2013-07-23 | | Release date: | 2014-05-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones.
Nature, 511, 2014
|
|
2JJC
 
 | | Hsp90 alpha ATPase domain with bound small molecule fragment | | Descriptor: | DIMETHYL SULFOXIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA, PYRIMIDIN-2-AMINE | | Authors: | Congreve, M, Chessari, G, Tisi, D, Woodhead, A.J. | | Deposit date: | 2008-03-31 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Recent Developments in Fragment-Based Drug Discovery.
J.Med.Chem., 51, 2008
|
|
