7B1S
 
 | | Crystal structure of the ethyl-coenzyme M reductase from Candidatus Ethanoperedens thermophilum at 0.994-A resolution | | Descriptor: | (2S)-2-{[(2S)-2-{[(2S)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wagner, T, Lemaire, O.N, Engilberge, S. | | Deposit date: | 2020-11-25 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.992 Å) | | Cite: | Crystal structure of a key enzyme for anaerobic ethane activation.
Science, 373, 2021
|
|
4BFX
 
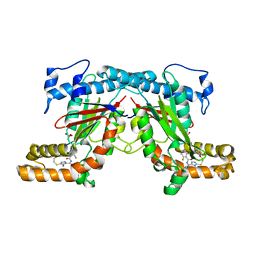 | | Crystal structure of Mycobacterium tuberculosis PanK in complex with a triazole inhibitory compound (1f) and phosphate | | Descriptor: | 2,6-difluoro-N-[1-(5-{[2-(4-fluorophenoxy)ethyl]sulfanyl}-4-methyl-4H-1,2,4-triazol-3-yl)ethyl]benzamide, PANTOTHENATE KINASE, PHOSPHATE ION | | Authors: | Bjorkelid, C, Bergfors, T, Jones, T.A. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Characterization of Compounds Inhibiting Mycobacterium Tuberculosis Pank
J.Biol.Chem., 288, 2013
|
|
5YQ7
 
 | | Cryo-EM structure of the RC-LH core complex from Roseiflexus castenholzii | | Descriptor: | 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, BACTERIOCHLOROPHYLL A, ... | | Authors: | Shi, Y, Xin, Y.Y, Niu, T.X, Wang, Q.Q, Niu, W.Q, Huang, X.J, Ding, W, Blankenship, R.E, Xu, X.L, Sun, F. | | Deposit date: | 2017-11-05 | | Release date: | 2018-05-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the RC-LH core complex from an early branching photosynthetic prokaryote.
Nat Commun, 9, 2018
|
|
5UQ0
 
 | |
7QNC
 
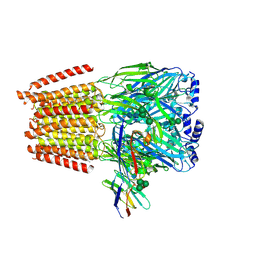 | | Cryo-EM structure of human full-length extrasynaptic alpha4beta3delta GABA(A)R in complex with THIP (gaboxadol), histamine and nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,5,6,7-tetrahydro-[1,2]oxazolo[5,4-c]pyridin-3-one, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
2XVS
 
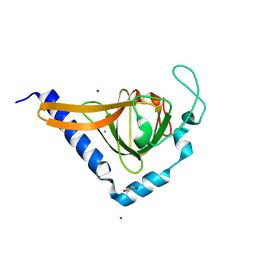 | | Crystal structure of human TTC5 (Strap) C-terminal OB domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IODIDE ION, ... | | Authors: | Adams, J, Pike, A.C.W, Maniam, S, Sharpe, T.D, Coutts, A.S, Knapp, S, La Thangue, B, Bullock, A.N. | | Deposit date: | 2010-10-31 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The P53 Cofactor Strap Exhibits an Unexpected Tpr Motif and Oligonucleotide-Binding (Ob)-Fold Structure.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
9JVX
 
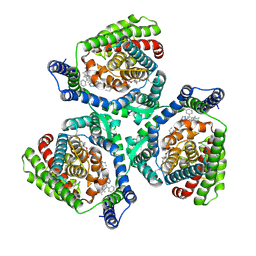 | | Overall structure of human EAAT2 bound with activator (GT949) | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 3-[(~{R})-(4-cyclohexylpiperazin-1-yl)-[1-(2-phenylethyl)-1,2,3,4-tetrazol-5-yl]methyl]-6-methoxy-1~{H}-quinolin-2-one, CHOLESTEROL, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Shi, Y, Huang, J, Zhou, Q. | | Deposit date: | 2024-10-09 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural study of human glutamate transporter EAAT2
Dian Zi Xian Wei Xue Bao, 2025
|
|
4AW8
 
 | | X-ray structure of ZinT from Salmonella enterica in complex with zinc ion and PEG | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Metal-binding protein ZinT, SODIUM ION, ... | | Authors: | Alaleona, F, Ilari, A, Battistoni, A, Petrarca, P, Chiancone, E. | | Deposit date: | 2012-06-01 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Salmonella Enterica Zint Structure, Zinc Affinity and Interaction with the High-Affinity Uptake Protein Znua Provide Insight Into the Management of Periplasmic Zinc.
Biochim.Biophys.Acta, 1840, 2014
|
|
4KNH
 
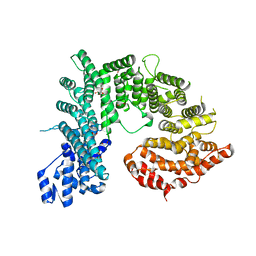 | | Structure of the Chaetomium thermophilum adaptor nucleoporin Nup192 N-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Nup192p, ... | | Authors: | Stuwe, T, Lin, D.H, Collins, L.N, Hoelz, A. | | Deposit date: | 2013-05-09 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Evidence for an evolutionary relationship between the large adaptor nucleoporin Nup192 and karyopherins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5DTT
 
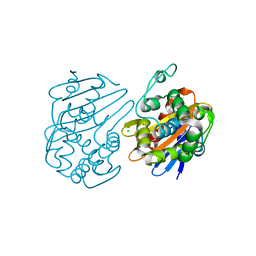 | | Fragments bound to the OXA-48 beta-lactamase: Compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 3-(1,3-thiazol-2-yl)benzoic acid, Beta-lactamase, ... | | Authors: | Lund, B.A, Christopeit, T, Leiros, H.-K.S. | | Deposit date: | 2015-09-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.10000539 Å) | | Cite: | Screening and Design of Inhibitor Scaffolds for the Antibiotic Resistance Oxacillinase-48 (OXA-48) through Surface Plasmon Resonance Screening.
J.Med.Chem., 59, 2016
|
|
7AOS
 
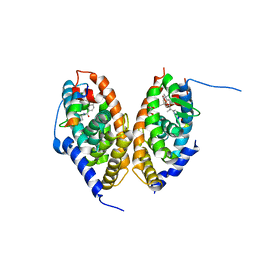 | | crystal structure of the RARalpha/RXRalpha ligand binding domain heterodimer in complex with a fragment of SRC1 coactivator | | Descriptor: | 4-{[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbonyl]amino}benzoic acid, 6-[1-(3,5,5,8,8-PENTAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)CYCLOPROPYL]PYRIDINE-3-CARBOXYLIC ACID, GLYCEROL, ... | | Authors: | le Maire, A, Guee, L, Bourguet, W. | | Deposit date: | 2020-10-15 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights into the Interaction of the Intrinsically Disordered Co-activator TIF2 with Retinoic Acid Receptor Heterodimer (RXR/RAR).
J.Mol.Biol., 433, 2021
|
|
6RBD
 
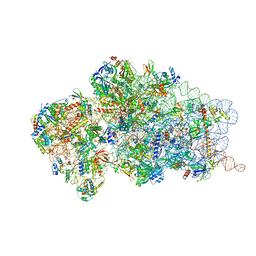 | | State 1 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
9NJI
 
 | | M298L Streptomyces coelicolor Laccase | | Descriptor: | COPPER (II) ION, Copper oxidase, GLYCINE, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2025-02-27 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unexpected effect of an axial ligand mutation in the type 1 copper center in small laccase: structure-based analyses and engineering to increase reduction potential and activity.
Chem Sci, 16, 2025
|
|
2WUG
 
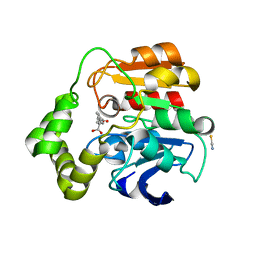 | | Crystal structure of S114A mutant of HsaD from Mycobacterium tuberculosis in complex with HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, ... | | Authors: | Lack, N.A, Yam, K.C, Lowe, E.D, Horsman, G.P, Owen, R.L, Sim, E, Eltis, L.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a C-C Hydrolase from Mycobacterium Tuberuclosis Involved in Cholesterol Metabolism.
J.Biol.Chem., 285, 2010
|
|
2XVN
 
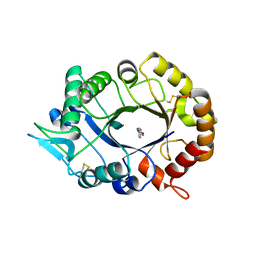 | | A. fumigatus chitinase A1 phenyl-methylguanylurea complex | | Descriptor: | 1-METHYL-3-(N-PHENYLCARBAMIMIDOYL)UREA, ASPERGILLUS FUMIGATUS CHITINASE A1, CHLORIDE ION | | Authors: | Rush, C.L, Schuttelkopf, A.W, Hurtado-Guerrero, R, Blair, D.E, Ibrahim, A.F.M, Desvergnes, S, Eggleston, I.M, van Aalten, D.M.F. | | Deposit date: | 2010-10-26 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Natural Product-Guided Discovery of a Fungal Chitinase Inhibitor.
Chem.Biol., 17, 2010
|
|
5V54
 
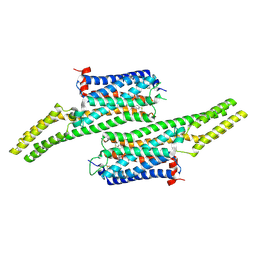 | | Crystal structure of 5-HT1B receptor in complex with methiothepin | | Descriptor: | 1-methyl-4-[(5~{S})-3-methylsulfanyl-5,6-dihydrobenzo[b][1]benzothiepin-5-yl]piperazine, 5-hydroxytryptamine receptor 1B,OB-1 fused 5-HT1b receptor,5-hydroxytryptamine receptor 1B | | Authors: | Yin, W.C, Zhou, X.E, Yang, D, de Waal, P, Wang, M.T, Dai, A, Cai, X, Huang, C.Y, Liu, P, Yin, Y, Liu, B, Caffrey, M, Melcher, K, Xu, Y, Wang, M.W, Xu, H.E, Jiang, Y. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | A common antagonistic mechanism for class A GPCRs revealed by the structure of the human 5-HT1B serotonin receptor bound to an antagonist
Cell Discov, 2018
|
|
4YMA
 
 | | Structure of the ligand-binding domain of GluA2 in complex with the antagonist CNG10109 | | Descriptor: | (3R)-3-(3-carboxy-5-hydroxyphenyl)-L-proline, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Moller, C, Tapken, D, Kastrup, J.S, Frydenvang, K. | | Deposit date: | 2015-03-06 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structure-Activity Relationship Study of Ionotropic Glutamate Receptor Antagonist (2S,3R)-3-(3-Carboxyphenyl)pyrrolidine-2-carboxylic Acid.
J.Med.Chem., 58, 2015
|
|
2WVA
 
 | | Structural insights into the pre-reaction state of pyruvate decarboxylase from Zymomonas mobilis | | Descriptor: | 2-{1-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-METHYL-1H-1,2,3-TRIAZOL-4-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, PYRUVATE DECARBOXYLASE, ... | | Authors: | Pei, X.Y, Erixon, K, Luisi, B.F, Leeper, F.J. | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into the Pre-Reaction State of Pyruvate Decarboxylase from Zymomonas Mobilis
Biochemistry, 49, 2010
|
|
1R3J
 
 | | potassium channel KcsA-Fab complex in high concentration of Tl+ | | Descriptor: | Antibody Fab fragment heavy chain, Antibody Fab fragment light chain, DIACYL GLYCEROL, ... | | Authors: | Zhou, Y, MacKinnon, R. | | Deposit date: | 2003-10-02 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The occupancy of ions in the K+ selectivity filter: Charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates
J.Mol.Biol., 333, 2003
|
|
5V71
 
 | | KRAS G12C in bound to quinazoline based switch II pocket (SWIIP) binder | | Descriptor: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Westover, K, Lu, J. | | Deposit date: | 2017-03-17 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.228 Å) | | Cite: | KRAS G12C Drug Development: Discrimination between Switch II Pocket Configurations Using Hydrogen/Deuterium-Exchange Mass Spectrometry.
Structure, 25, 2017
|
|
2WHJ
 
 | | Understanding how diverse mannanases recognise heterogeneous substrates | | Descriptor: | ACETATE ION, BETA-MANNANASE, GLYCEROL, ... | | Authors: | Tailford, L.E, Ducros, V.M.A, Flint, J.E, Roberts, S.M, Morland, C, Zechel, D.L, Smith, N, Bjornvad, M.E, Borchert, T.V, Wilson, K.S, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2009-05-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Understanding How Diverse -Mannanases Recognise Heterogeneous Substrates.
Biochemistry, 48, 2009
|
|
4YMB
 
 | | Structure of the ligand-binding domain of GluK1 in complex with the antagonist CNG10111 | | Descriptor: | (3R,4S)-3-(3-carboxyphenyl)-4-propyl-L-proline, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Moller, C, Tapken, D, Kastrup, J.S, Frydenvang, K. | | Deposit date: | 2015-03-06 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-Activity Relationship Study of Ionotropic Glutamate Receptor Antagonist (2S,3R)-3-(3-Carboxyphenyl)pyrrolidine-2-carboxylic Acid.
J.Med.Chem., 58, 2015
|
|
4FNZ
 
 | | Crystal structure of human anaplastic lymphoma kinase in complex with piperidine-carboxamide inhibitor 2 | | Descriptor: | (3S)-N-[3-(trifluoromethoxy)benzyl]-1-{2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}piperidine-3-carboxamide, ALK tyrosine kinase receptor | | Authors: | Whittington, D.A, Epstein, L.F, Chen, H. | | Deposit date: | 2012-06-20 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The R1275Q Neuroblastoma Mutant and Certain ATP-competitive Inhibitors Stabilize Alternative Activation Loop Conformations of Anaplastic Lymphoma Kinase.
J.Biol.Chem., 287, 2012
|
|
5VC3
 
 | | CRYSTAL STRUCTURE OF HUMAN WEE1 KINASE DOMAIN IN COMPLEX WITH BOSUTINIB | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-03-30 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
3UB0
 
 | | Crystal structure of the nonstructural protein 7 and 8 complex of Feline Coronavirus | | Descriptor: | Non-structural protein 6, nsp6,, Non-structural protein 7, ... | | Authors: | Xiao, Y, Hilgenfeld, R, Ma, Q. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity.
J.Virol., 86, 2012
|
|
