7YV5
 
 | |
7Z8Z
 
 | | Crystal structure of the MEILB2-BRME1 2:2 core complex | | Descriptor: | Break repair meiotic recombinase recruitment factor 1, Heat shock factor 2-binding protein | | Authors: | Gurusaran, M, Davies, O.R. | | Deposit date: | 2022-03-19 | | Release date: | 2023-09-20 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | MEILB2-BRME1 forms a V-shaped DNA clamp upon BRCA2-binding in meiotic recombination.
Nat Commun, 15, 2024
|
|
7SXP
 
 | |
8CZO
 
 | | Cryo-EM structure of BCL10 CARD - MALT1 DD filament | | Descriptor: | B-cell lymphoma/leukemia 10, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | David, L, Wu, H. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | BCL10 Mutations Define Distinct Dependencies Guiding Precision Therapy for DLBCL.
Cancer Discov, 12, 2022
|
|
8CZD
 
 | | Cryo-EM structure of BCL10 R58Q filament | | Descriptor: | B-cell lymphoma/leukemia 10 | | Authors: | David, L, Wu, H. | | Deposit date: | 2022-05-24 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | BCL10 Mutations Define Distinct Dependencies Guiding Precision Therapy for DLBCL.
Cancer Discov, 12, 2022
|
|
6RN8
 
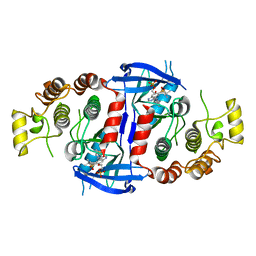 | | RIP2 Kinase Catalytic Domain complex with 2(4[(1,3benzothiazol5yl)amino]6(2methylpropane2sulfonyl)quinazolin7yl)oxy)ethyl phosphate | | Descriptor: | 2-[4-(1,3-benzothiazol-5-ylamino)-6-~{tert}-butylsulfonyl-quinazolin-7-yl]oxyethyl dihydrogen phosphate, CALCIUM ION, Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Convery, M.A, Haile, P.A. | | Deposit date: | 2019-05-08 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of a First-in-Class Receptor Interacting Protein 2 (RIP2) Kinase Specific Clinical Candidate, 2-((4-(Benzo[d]thiazol-5-ylamino)-6-(tert-butylsulfonyl)quinazolin-7-yl)oxy)ethyl Dihydrogen Phosphate, for the Treatment of Inflammatory Diseases.
J.Med.Chem., 62, 2019
|
|
5K8G
 
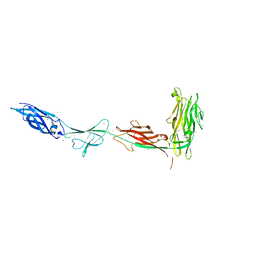 | |
6RNA
 
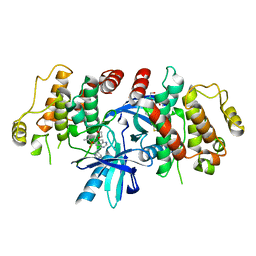 | | RIP2 Kinase Catalytic Domain complex with 2({4[(1,3benzothiazol5yl)amino]6(2methylpropane2sulfonyl)quinazolin7yl}oxy)ethan1ol | | Descriptor: | 2-[4-(1,3-benzothiazol-5-ylamino)-6-~{tert}-butylsulfonyl-quinazolin-7-yl]oxyethanol, Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Convery, M.A, Haile, P.A. | | Deposit date: | 2019-05-08 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Discovery of a First-in-Class Receptor Interacting Protein 2 (RIP2) Kinase Specific Clinical Candidate, 2-((4-(Benzo[d]thiazol-5-ylamino)-6-(tert-butylsulfonyl)quinazolin-7-yl)oxy)ethyl Dihydrogen Phosphate, for the Treatment of Inflammatory Diseases.
J.Med.Chem., 62, 2019
|
|
1RFI
 
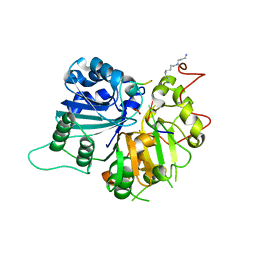 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, pentapeptide KLNYK, and tetranucleotide AGTC | | Descriptor: | 5'-D(*AP*GP*TP*C)-3', SPERMINE, Topoisomerase I-Derived Peptide, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
3P2X
 
 | | Insulin fibrillation is the Janus face of induced fit. A chiaral clamp stabilizes the native state at the expense of activity | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Hua, Q.X, Wan, Z.L, Huang, K, Hu, S.Q, Phillip, N.F, Jia, W.H, Whittingham, J, Dodson, G.G, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2010-10-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity
To be Published
|
|
3P33
 
 | | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Hua, Q.X, Wan, Z.L, Huang, K, Hu, S.Q, Phillip, N.F, Jia, W.H, Whittingham, J, Dodson, G.G, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2010-10-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity
To be Published
|
|
8D73
 
 | | Crystal Structure of EGFR LRTM with compound 7 | | Descriptor: | (3S,4R)-3-fluoro-1-(4-{[4-(methylamino)-1-(propan-2-yl)pyrido[3,4-d]pyridazin-7-yl]amino}pyrimidin-2-yl)piperidin-4-ol, Epidermal growth factor receptor, GLYCEROL | | Authors: | Kim, J.L. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery of BLU-945, a Reversible, Potent, and Wild-Type-Sparing Next-Generation EGFR Mutant Inhibitor for Treatment-Resistant Non-Small-Cell Lung Cancer.
J.Med.Chem., 65, 2022
|
|
6XLC
 
 | | Full-length Hsc82 bound to AMPPNP | | Descriptor: | ATP-dependent molecular chaperone HSC82, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Liu, Y.X, Sun, M, Myasnikov, A.G, Elnatan, D, Agard, D.A. | | Deposit date: | 2020-06-28 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Full-length Hsc82 bound to AMPPNP
To Be Published
|
|
8D76
 
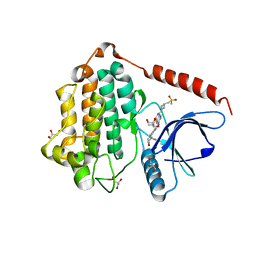 | | Crystal Structure of EGFR LRTM with compound 24 | | Descriptor: | (3S,4R)-3-fluoro-1-(4-{[8-{3-[(methanesulfonyl)methyl]azetidin-1-yl}-5-(propan-2-yl)-2,7-naphthyridin-3-yl]amino}pyrimidin-2-yl)-3-methylpiperidin-4-ol, Epidermal growth factor receptor, GLYCEROL | | Authors: | Kim, J.L. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of BLU-945, a Reversible, Potent, and Wild-Type-Sparing Next-Generation EGFR Mutant Inhibitor for Treatment-Resistant Non-Small-Cell Lung Cancer.
J.Med.Chem., 65, 2022
|
|
6J8Y
 
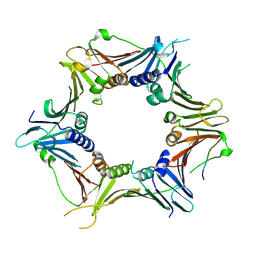 | | Crystal structure of the human RAD9-HUS1-RAD1-RHINO complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Hara, K, Iida, N, Sakurai, H, Hashimoto, H. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RAD9-RAD1-HUS1 checkpoint clamp bound to RHINO sheds light on the other side of the DNA clamp.
J.Biol.Chem., 295, 2020
|
|
830C
 
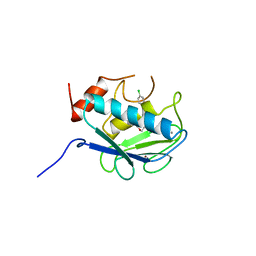 | | COLLAGENASE-3 (MMP-13) COMPLEXED TO A SULPHONE-BASED HYDROXAMIC ACID | | Descriptor: | 4-[4-(4-CHLORO-PHENOXY)-BENZENESULFONYLMETHYL]-TETRAHYDRO-PYRAN-4-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, MMP-13, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
7U6U
 
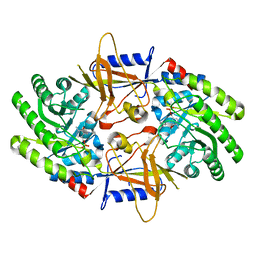 | | Structure of an intellectual disability-associated ornithine decarboxylase variant G84R in complex with PLP | | Descriptor: | Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Schultz, C.R, Powell, K.S, Henrickson, A, Lamp, J, Brunzelle, J.S, Demeler, B, Vega, I.E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2022-03-06 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Enzymatic Activity of an Intellectual Disability-Associated Ornithine Decarboxylase Variant, G84R.
Acs Omega, 7, 2022
|
|
7U6P
 
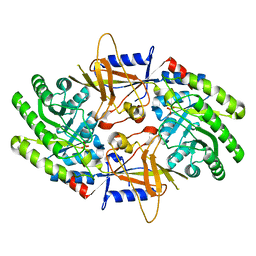 | | Structure of an intellectual disability-associated ornithine decarboxylase variant G84R | | Descriptor: | Ornithine decarboxylase, PHOSPHATE ION | | Authors: | Zhou, X.E, Schultz, C.R, Powell, K.S, Henrickson, A, Lamp, J, Brunzelle, J.S, Demeler, B, Vega, I.E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Enzymatic Activity of an Intellectual Disability-Associated Ornithine Decarboxylase Variant, G84R.
Acs Omega, 7, 2022
|
|
3PYZ
 
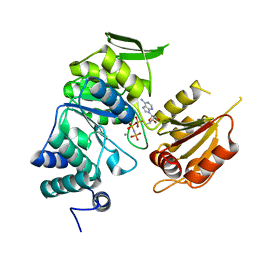 | | Crystal structure of bifunctional folylpolyglutamate synthase/dihydrofolate synthase complexed with AMPPNP and Mn ion from Yersinia pestis c092 | | Descriptor: | Bifunctional folylpolyglutamate synthase/dihydrofolate synthase, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of bifunctional folylpolyglutamate synthase/dihydrofolate synthase complexed with AMPpnp and Mn ion from Yersinia pestis c092
TO BE PUBLISHED
|
|
3QCT
 
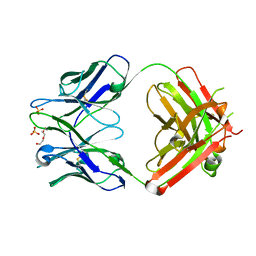 | | Crystal structure of the humanized apo LT3015 anti-lysophosphatidic acid antibody Fab fragment | | Descriptor: | LT3015 antibody Fab fragment, heavy chain, light chain, ... | | Authors: | Fleming, J.K, Wojciak, J.M, Campbell, M.-A, Huxford, T. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1493 Å) | | Cite: | Biochemical and structural characterization of lysophosphatidic Acid binding by a humanized monoclonal antibody.
J.Mol.Biol., 408, 2011
|
|
7CMP
 
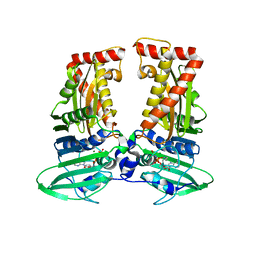 | | parE in complex with AMPPNP | | Descriptor: | DNA topoisomerase 4 subunit B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Jung, H.Y, Heo, Y.-S. | | Deposit date: | 2020-07-28 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | Crystal structure of parE in complex with AMPPNP
To Be Published
|
|
3QCY
 
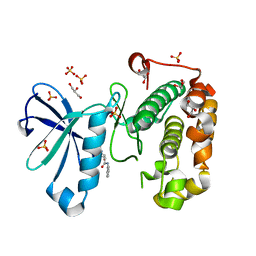 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 4-[2-Amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-N-phenyl-2-morpholinecarboxamide | | Descriptor: | (2S)-4-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-N-phenylmorpholine-2-carboxamide, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QCX
 
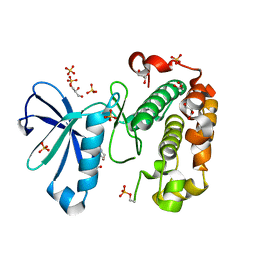 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-{2-Amino-6-[(3R)-3-methyl-4-morpholinyl]-4-pyrimidinyl}-1H-indazol-3-amine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-{2-amino-6-[(3R)-3-methylmorpholin-4-yl]pyrimidin-4-yl}-2H-indazol-3-amine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
8ORV
 
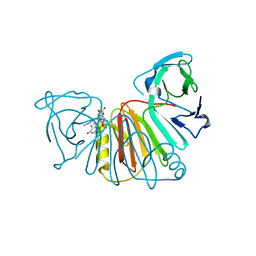 | | Crystal structure of monkeypox virus poxin in complex with the STING agonist MD1203 | | Descriptor: | 9-[(1~{S},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, MPXVgp165 | | Authors: | Duchoslav, V, Klima, M, Boura, E. | | Deposit date: | 2023-04-17 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
8P45
 
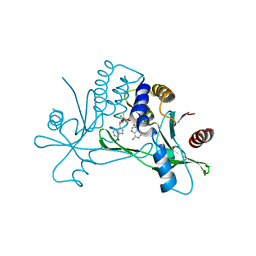 | | Crystal structure of human STING in complex with the agonist MD1202D | | Descriptor: | 9-[(1~{S},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2023-05-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
