7A78
 
 | | Crystal structure of RXR beta LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
6SAF
 
 | | The Fk1 domain of FKBP51 in complex with (S)-(R)-3-(3,4-dimethoxyphenyl)-1-(3-(2-morpholinoethoxy)phenyl)propyl 1-((1R,4aR,8aR)-4-oxodecahydronaphthalene-1-carbonyl)piperidine-2-carboxylate | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, [(1~{R})-3-(3,4-dimethoxyphenyl)-1-[3-(2-morpholin-4-ylethoxy)phenyl]propyl] (2~{S})-1-[[(1~{R},4~{a}~{R},8~{a}~{R})-4-oxidanylidene-2,3,4~{a},5,6,7,8,8~{a}-octahydro-1~{H}-naphthalen-1-yl]carbonyl]piperidine-2-carboxylate | | Authors: | Feng, X, Sippel, C, Knaup, F, Bracher, A, Staibano, S, Romano, M.F, Hausch, F. | | Deposit date: | 2019-07-16 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Novel Decalin-Based Bicyclic Scaffold for FKBP51-Selective Ligands.
J.Med.Chem., 63, 2020
|
|
6BZ8
 
 | | Thermus thermophilus 70S containing 16S G347U point mutation and near-cognate ASL Leucine in A site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2017-12-22 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding.
Nucleic Acids Res., 47, 2019
|
|
7UFV
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-6766 | | Descriptor: | (3P)-N-[(1S)-3-amino-1-(3-chlorophenyl)-3-oxopropyl]-3-(2-fluorophenyl)-1H-pyrazole-4-carboxamide, DDB1- and CUL4-associated factor 1, UNKNOWN ATOM OR ION | | Authors: | Kimani, S, Li, A, Li, Y, Dong, A, Hutchinson, A, Seitova, A, Wilson, B, Al-Awar, R, Vedadi, M, Brown, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nanomolar DCAF1 Small Molecule Ligands.
J.Med.Chem., 66, 2023
|
|
6BZ6
 
 | | Thermus thermophilus 70S complex containing 16S G347U ram mutation and empty A site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2017-12-22 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding.
Nucleic Acids Res., 47, 2019
|
|
8P58
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 500 micromolar X77 enantiomer R. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P86
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 mM MG-132, from an "old" crystal. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-31 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
6STI
 
 | | Crystal structure of RXRalpha LBD in complex with LG 100754 and a coactivator peptide | | Descriptor: | (2E,4E,6Z)-3-methyl-7-(5,5,8,8-tetramethyl-3-propoxy-5,6,7,8-tetrahydronaphthalen-2-yl)octa-2,4,6-trienoic acid, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | le Maire, A, Teyssier, C, Germain, P, Bourguet, W. | | Deposit date: | 2019-09-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Regulation of RXR-RAR Heterodimers by RXR- and RAR-Specific Ligands and Their Combinations.
Cells, 8, 2019
|
|
6BUW
 
 | | Thermus thermophilus 70S complex containing 16S G299A ram mutation and empty A site. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2017-12-11 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding.
Nucleic Acids Res., 47, 2019
|
|
6BZ7
 
 | | Thermus thermophilus 70S containing 16S G299A point mutation and near-cognate ASL Leucine in A site. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2017-12-22 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding.
Nucleic Acids Res., 47, 2019
|
|
6SSQ
 
 | | Crystal structure of RARbeta LBD in complex with LG 100754 | | Descriptor: | (2E,4E,6Z)-3-methyl-7-(5,5,8,8-tetramethyl-3-propoxy-5,6,7,8-tetrahydronaphthalen-2-yl)octa-2,4,6-trienoic acid, CITRATE ANION, GLYCEROL, ... | | Authors: | le Maire, A, Teyssier, C, Germain, P, Bourguet, W. | | Deposit date: | 2019-09-09 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulation of RXR-RAR Heterodimers by RXR- and RAR-Specific Ligands and Their Combinations.
Cells, 8, 2019
|
|
7O7G
 
 | | Crystal structure of the Shewanella oneidensis MR1 MtrC mutant H561M | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Edwards, M.J, van Wonderen, J.H, Newton-Payne, S.E, Butt, J.N, Clarke, T.A. | | Deposit date: | 2021-04-13 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nanosecond heme-to-heme electron transfer rates in a multiheme cytochrome nanowire reported by a spectrally unique His/Met-ligated heme.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8FLK
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2 | | Descriptor: | 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
8FLM
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53 | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, ... | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
7XKE
 
 | | Cryo-EM structure of DHEA-ADGRG2-FL-Gs complex | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKF
 
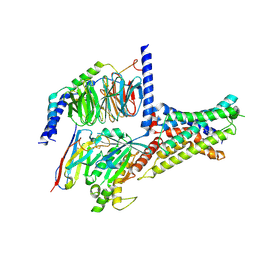 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex at lower state | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKD
 
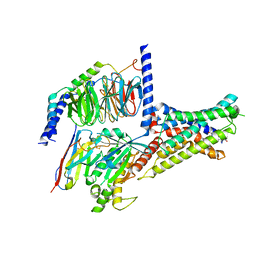 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
3D6D
 
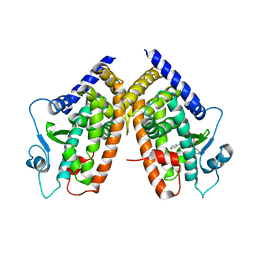 | | Crystal Structure of the complex between PPARgamma LBD and the LT175(R-enantiomer) | | Descriptor: | (2S)-2-(biphenyl-4-yloxy)-3-phenylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R. | | Deposit date: | 2008-05-19 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Peroxisome Proliferator-Activated Receptor gamma (PPARgamma) Ligand Binding Domain Complexed with a Novel Partial Agonist: A New Region of the Hydrophobic Pocket Could Be Exploited for Drug Design
J.Med.Chem., 51, 2008
|
|
3DT3
 
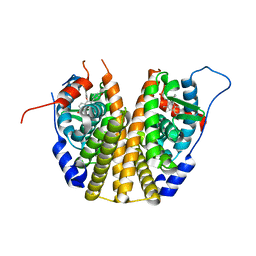 | | Human Estrogen receptor alpha LBD with GW368 | | Descriptor: | 5-(4-hydroxyphenoxy)-6-(3-hydroxyphenyl)-7-methylnaphthalen-2-ol, Estrogen receptor | | Authors: | Williams, S.P, Miller, A.B. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis of 3-alkyl naphthalenes as novel estrogen receptor ligands.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4V4C
 
 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ACETATE ION, CALCIUM ION, ... | | Authors: | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | Deposit date: | 2004-06-02 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
4V4E
 
 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici complexed with inhibitor 1,2,4,5-tetrahydroxy-benzene | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, BENZENE-1,2,4,5-TETROL, CALCIUM ION, ... | | Authors: | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | Deposit date: | 2004-06-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
6YMM
 
 | |
7O50
 
 | | Crystal structure of human legumain in complex with Gly-Ser-Asn peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLY-SER-ASN, ... | | Authors: | Dall, E, Brandstetter, H. | | Deposit date: | 2021-04-07 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Peptide Ligase Activity of Human Legumain Depends on Fold Stabilization and Balanced Substrate Affinities.
Acs Catalysis, 11, 2021
|
|
7WVF
 
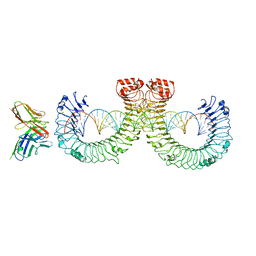 | | ectoTLR3-mAb12-poly(I:C) complex | | Descriptor: | RNA (46-MER), Toll-like receptor 3, mAb12 | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-10 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7WVJ
 
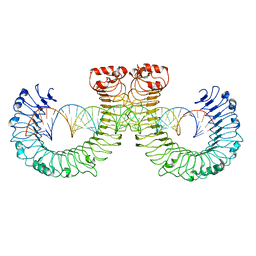 | | NT-mut(K117D,K139D,K145D) TLR3 -poly I:C complex | | Descriptor: | RNA (46-MER), Toll-like receptor 3 | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-10 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
