8K2I
 
 | | Crystal structure of Group 2 Oligosaccharide/Monosaccharide-releasing beta-N-acetylhexosaminidase NgaAt from Arabidopsis thaliana in complex with GlcNAc-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sumida, T, Fushinobu, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Genetic and functional diversity of beta-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis.
Nat Commun, 15, 2024
|
|
8DAF
 
 | | Human SF-1 LBD bound to synthetic agonist 6N-10CA and bacterial phospholipid | | Descriptor: | 10-[(3aR,6S,6aR)-3-phenyl-3a-(1-phenylethenyl)-6-(sulfamoylamino)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid (non-preferred name), DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor coactivator 2, ... | | Authors: | D'Agostino, E.H, Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparison of activity, structure, and dynamics of SF-1 and LRH-1 complexed with small molecule modulators.
J.Biol.Chem., 299, 2023
|
|
8K57
 
 | |
7TVD
 
 | |
8D1X
 
 | |
8JRL
 
 | | Crystal structure of P450 TleB with an indole alkaloid | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-(4-fluoranyl-1~{H}-indol-3-yl)-3-oxidanyl-propan-2-yl]-3-methyl-2-sulfanyl-butanamide, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, J, Yan, W.P. | | Deposit date: | 2023-06-17 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Regulation of P450 TleB catalytic flow for the synthesis of sulfur-containing indole alkaloids by substrate structure-directed strategy. and protein engineering.
Sci China Chem, 66, 2023
|
|
8D0B
 
 | | Human CST-DNA polymerase alpha/primase preinitiation complex bound to 4xTEL-foldback template | | Descriptor: | CST complex subunit CTC1, CST complex subunit STN1, CST complex subunit TEN1, ... | | Authors: | He, Q, Lin, X, Chavez, B.L, Agrawal, S, Lusk, B.L, Lim, C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Structures of the human CST-Pol alpha-primase complex bound to telomere templates.
Nature, 608, 2022
|
|
8K3C
 
 | | Nipah virus Attachment glycoprotein with 41-6 antibody fragment | | Descriptor: | Glycoprotein G, Heavy chain of 41-6 Fab fragments, Light chain of 41-6 Fab fragment | | Authors: | Sun, M.M. | | Deposit date: | 2023-07-15 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Potent human neutralizing antibodies against Nipah virus derived from two ancestral antibody heavy chains.
Nat Commun, 15, 2024
|
|
8JJA
 
 | | SP1746 in complex with acetate ions | | Descriptor: | ACETATE ION, FE (III) ION, bis(5'-nucleosyl)-tetraphosphatase (symmetrical) | | Authors: | Jin, Y, Niu, L, Ke, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 32, 2024
|
|
8D0K
 
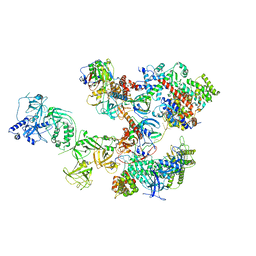 | | Human CST-DNA polymerase alpha/primase preinitiation complex bound to 4xTEL-foldback template - PRIM2C advanced PIC | | Descriptor: | CST complex subunit CTC1, CST complex subunit STN1, CST complex subunit TEN1, ... | | Authors: | He, Q, Lin, X, Chavez, B.L, Agrawal, S, Lusk, B.L, Lim, C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structures of the human CST-Pol alpha-primase complex bound to telomere templates.
Nature, 608, 2022
|
|
8K8T
 
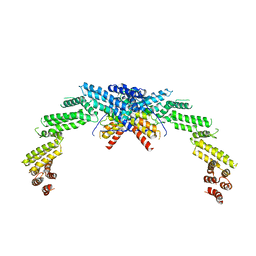 | | Structure of CUL3-RBX1-KLHL22 complex | | Descriptor: | Cullin-3, Kelch-like protein 22 | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
5YNG
 
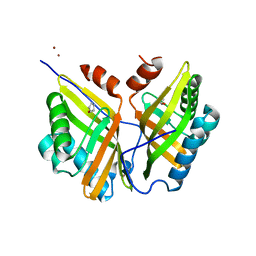 | | Crystal structure of SZ348 in complex with cyclopentene oxide | | Descriptor: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase, NICKEL (II) ION, ... | | Authors: | Wu, L, Sun, Z.T, Reetz, M.T, Zhou, J.H. | | Deposit date: | 2017-10-24 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structural and Computational Insight into the Catalytic Mechanism of Limonene Epoxide Hydrolase Mutants in Stereoselective Transformations.
J. Am. Chem. Soc., 140, 2018
|
|
7TF2
 
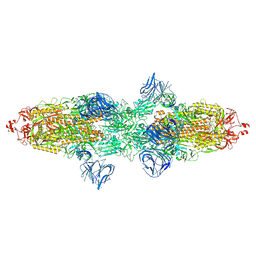 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) Q484I spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
7TF5
 
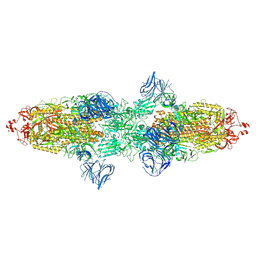 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
5YLA
 
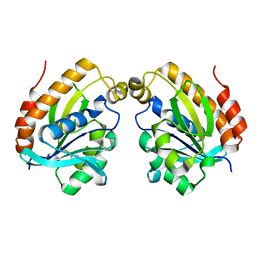 | |
5YLH
 
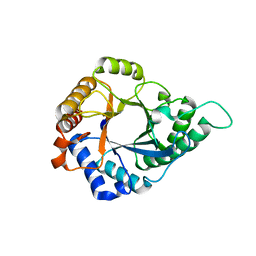 | | Structure of GH113 beta-1,4-mannanase | | Descriptor: | beta-1,4-mannanase | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase from Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
8JUU
 
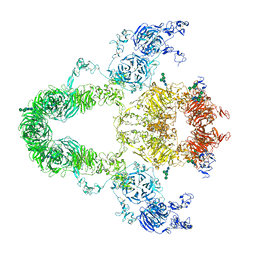 | | rat megalin | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-27 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8CTO
 
 | |
7U6Z
 
 | | Pertussis toxin E129D NAD | | Descriptor: | IODIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pertussis toxin subunit 1 | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2022-03-06 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.30002 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
8CUN
 
 | |
8CWA
 
 | |
8D9O
 
 | | De Novo Photosynthetic Reaction Center Protein in Apo-State | | Descriptor: | CADMIUM ION, Reaction Center Maquette | | Authors: | Ennist, N.M, Stayrook, S.E, Dutton, P.L, Moser, C.C. | | Deposit date: | 2022-06-10 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Rational design of photosynthetic reaction center protein maquettes.
Front Mol Biosci, 9, 2022
|
|
5YGI
 
 | |
8D07
 
 | | Hallucinated C3 protein assembly HALC3_109 | | Descriptor: | HALC3_109 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8JUT
 
 | | rat megalin RAP complex | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-27 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
