2EOW
 
 | | Solution structure of the C2H2 type zinc finger (region 368-400) of human Zinc finger protein 347 | | Descriptor: | ZINC ION, Zinc finger protein 347 | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 368-400) of human Zinc finger protein 347
To be Published
|
|
2EMC
 
 | | Solution structure of the C2H2 type zinc finger (region 641-673) of human Zinc finger protein 473 | | Descriptor: | ZINC ION, Zinc finger protein 473 | | Authors: | Tomizawa, T, Tochio, N, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 641-673) of human Zinc finger protein 473
To be Published
|
|
2EMP
 
 | | Solution structure of the C2H2 type zinc finger (region 536-568) of human Zinc finger protein 347 | | Descriptor: | ZINC ION, Zinc finger protein 347 | | Authors: | Tomizawa, T, Tochio, N, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 536-568) of human Zinc finger protein 347
To be Published
|
|
2EN9
 
 | | Solution structure of the C2H2 type zinc finger (region 415-447) of human Zinc finger protein 28 homolog | | Descriptor: | ZINC ION, Zinc finger protein 28 homolog | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 415-447) of human Zinc finger protein 28 homolog
To be Published
|
|
2EM2
 
 | | Solution structure of the C2H2 type zinc finger (region 584-616) of human Zinc finger protein 28 homolog | | Descriptor: | ZINC ION, Zinc finger protein 28 homolog | | Authors: | Tomizawa, T, Tochio, N, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 584-616) of human Zinc finger protein 28 homolog
To be Published
|
|
2EMH
 
 | | Solution structure of the C2H2 type zinc finger (region 491-523) of human Zinc finger protein 484 | | Descriptor: | ZINC ION, Zinc finger protein 484 | | Authors: | Tomizawa, T, Tochio, N, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 491-523) of human Zinc finger protein 484
To be Published
|
|
2EN1
 
 | | Solution structure of the C2H2 type zinc finger (region 563-595) of human Zinc finger protein 224 | | Descriptor: | ZINC ION, Zinc finger protein 224 | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 563-595) of human Zinc finger protein 224
To be Published
|
|
8FFZ
 
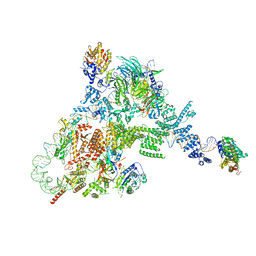 | | TFIIIA-TFIIIC-Brf1-TBP complex bound to 5S rRNA gene | | Descriptor: | DNA (151-MER), Transcription factor IIIA, Transcription factor IIIB 70 kDa subunit,TATA-box-binding protein, ... | | Authors: | Talyzina, A, He, Y. | | Deposit date: | 2022-12-11 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of TFIIIC-dependent RNA polymerase III transcription initiation.
Mol.Cell, 83, 2023
|
|
7LPS
 
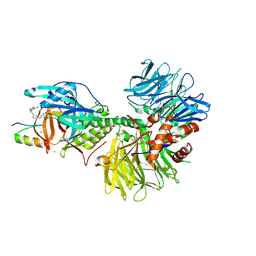 | | Crystal structure of DDB1-CRBN-ALV1 complex bound to Helios (IKZF2 ZF2) | | Descriptor: | 3-[3-[[1-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2,5-bis(oxidanylidene)pyrrol-3-yl]amino]phenyl]-~{N}-(3-chloranyl-4-methyl-phenyl)propanamide, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, Fischer, E.S. | | Deposit date: | 2021-02-12 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Acute pharmacological degradation of Helios destabilizes regulatory T cells.
Nat.Chem.Biol., 17, 2021
|
|
6QNX
 
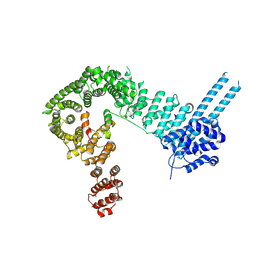 | | Structure of the SA2/SCC1/CTCF complex | | Descriptor: | Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog, Transcriptional repressor CTCF | | Authors: | Li, Y, Muir, K.W, Panne, D. | | Deposit date: | 2019-02-12 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for cohesin-CTCF-anchored loops.
Nature, 578, 2020
|
|
7U8F
 
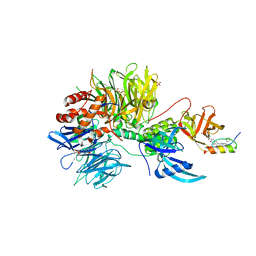 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2) and the molecular glue DKY709 | | Descriptor: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA damage-binding protein 1, ... | | Authors: | Ma, X, Ornelas, E, Clifton, M.C. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
7YSF
 
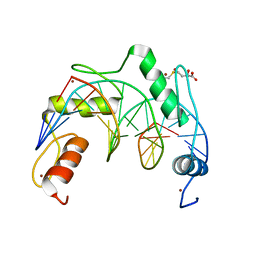 | | Crystal structure of ZNF524 ZF1-4 in complex with telomeric DNA | | Descriptor: | D-MALATE, DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*CP*C)-3'), ... | | Authors: | Li, F.D, Xu, Z.Y. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-16 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ZNF524 directly interacts with telomeric DNA and supports telomere integrity.
Nat Commun, 14, 2023
|
|
7W1M
 
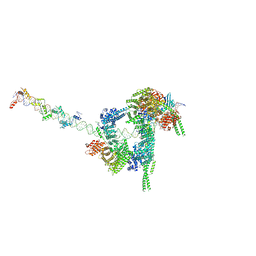 | | Cryo-EM structure of human cohesin-CTCF-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cohesin subunit SA-1, ... | | Authors: | Shi, Z.B, Bai, X.C, Yu, H. | | Deposit date: | 2021-11-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | CTCF and R-loops are boundaries of cohesin-mediated DNA looping.
Mol.Cell, 83, 2023
|
|
7BQV
 
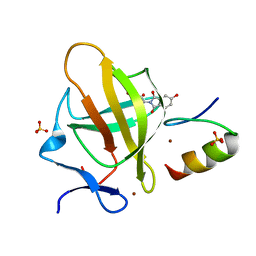 | | Cereblon in complex with SALL4 and (S)-5-hydroxythalidomide | | Descriptor: | 2-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-5-oxidanyl-isoindole-1,3-dione, Protein cereblon, SULFATE ION, ... | | Authors: | Furihata, H, Miyauchi, Y, Asano, A, Tanokura, M, Miyakawa, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural bases of IMiD selectivity that emerges by 5-hydroxythalidomide.
Nat Commun, 11, 2020
|
|
7BQU
 
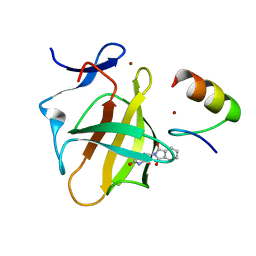 | | Cereblon in complex with SALL4 and (S)-thalidomide | | Descriptor: | Protein cereblon, S-Thalidomide, Sal-like protein 4, ... | | Authors: | Furihata, H, Miyauchi, Y, Asano, A, Tanokura, M, Miyakawa, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural bases of IMiD selectivity that emerges by 5-hydroxythalidomide.
Nat Commun, 11, 2020
|
|
6SKG
 
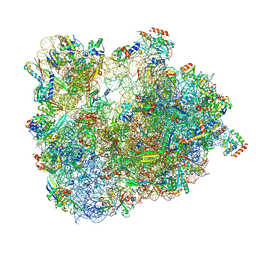 | | Cryo-EM Structure of T. kodakarensis 70S ribosome in TkNat10 deleted strain | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Sas-Chen, A, Thomas, J.M, Santangelo, T, Meier, J.L, Schwartz, S, Shalev-Benami, M. | | Deposit date: | 2019-08-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping.
Nature, 583, 2020
|
|
3W5K
 
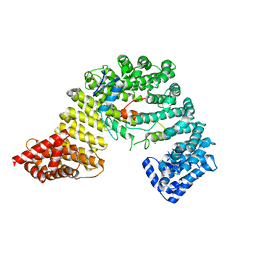 | | Crystal structure of Snail1 and importin beta complex | | Descriptor: | Importin subunit beta-1, ZINC ION, Zinc finger protein SNAI1 | | Authors: | Choi, S, Yamashita, E, Yasuhara, N, Song, J, Son, S.Y, Won, Y.H, Shin, Y.S, Sekimoto, T, Park, I.Y, Yoneda, Y, Lee, S.J. | | Deposit date: | 2013-01-30 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the selective nuclear import of the C2H2 zinc-finger protein Snail by importin beta.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3ZMS
 
 | | LSD1-CoREST in complex with INSM1 peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, INSULINOMA-ASSOCIATED PROTEIN 1, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
1LLM
 
 | |
8D80
 
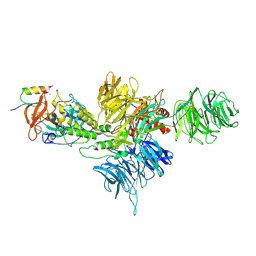 | | Cereblon~DDB1 bound to Iberdomide and Ikaros ZF1-2-3 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, DNA-binding protein Ikaros, ... | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8DEY
 
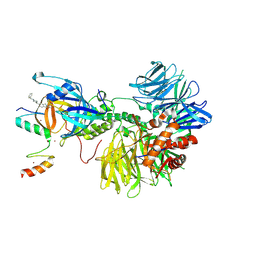 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2,3) and the molecular glue DKY709 | | Descriptor: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Ma, X, Ornelas, E, Clifton, M.C. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
8D7Z
 
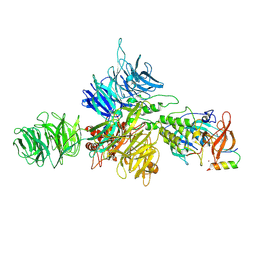 | | Cereblon-DDB1 bound to CC-92480 and Ikaros ZF1-2-3 | | Descriptor: | DNA damage-binding protein 1, DNA-binding protein Ikaros, Mezigdomide, ... | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
6H0F
 
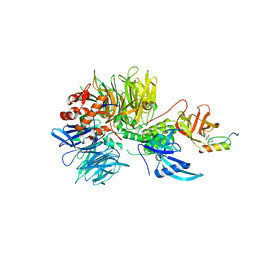 | | Structure of DDB1-CRBN-pomalidomide complex bound to IKZF1(ZF2) | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, DNA-binding protein Ikaros, Protein cereblon, ... | | Authors: | Petzold, G, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
6H0G
 
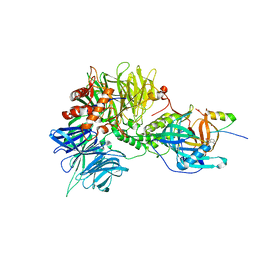 | | Structure of the DDB1-CRBN-pomalidomide complex bound to ZNF692(ZF4) | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DDB1 (DNA damage binding protein 1),DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Bunker, R.D, Petzold, G, Thoma, N.H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
6UML
 
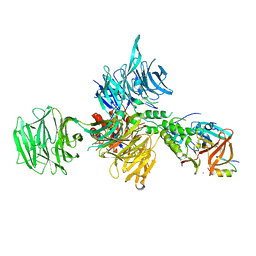 | | Structural Basis for Thalidomide Teratogenicity Revealed by the Cereblon-DDB1-SALL4-Pomalidomide Complex | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Clayton, T.L, Matyskiela, M.E, Pagarigan, B.E, Tran, E.T, Chamberlain, P.P. | | Deposit date: | 2019-10-09 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Crystal structure of the SALL4-pomalidomide-cereblon-DDB1 complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
