5D7N
 
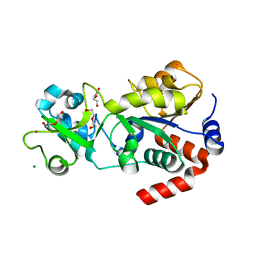 | | Crystal structure of human Sirt3 at an improved resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-08-14 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Seeding for sirtuins: microseed matrix seeding to obtain crystals of human Sirt3 and Sirt2 suitable for soaking.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2H4F
 
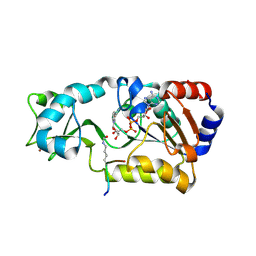 | | Sir2-p53 peptide-NAD+ | | Descriptor: | Cellular tumor antigen p53, NAD-dependent deacetylase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hoff, K.G, Avalos, J.L, Sens, K, Wolberger, C. | | Deposit date: | 2006-05-24 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the Sirtuin Mechanism from Ternary Complexes Containing NAD(+) and Acetylated Peptide.
Structure, 14, 2006
|
|
2H2I
 
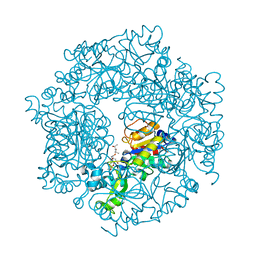 | | The Structural basis of Sirtuin Substrate Affinity | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, NAD-dependent deacetylase, ZINC ION | | Authors: | Cosgrove, M.S, Wolberger, C. | | Deposit date: | 2006-05-18 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of sirtuin substrate affinity
Biochemistry, 45, 2006
|
|
2H2G
 
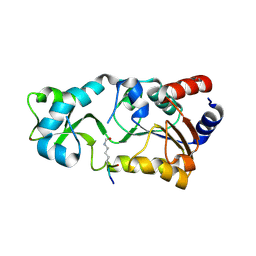 | |
2H4J
 
 | | Sir2-deacetylated peptide (from enzymatic turnover in crystal) | | Descriptor: | 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, Cellular tumor antigen p53, NAD-dependent deacetylase, ... | | Authors: | Hoff, K.G, Avalos, J.L, Sens, K, Wolberger, C. | | Deposit date: | 2006-05-24 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the Sirtuin Mechanism from Ternary Complexes Containing NAD(+) and Acetylated Peptide.
Structure, 14, 2006
|
|
2H2D
 
 | |
2H2F
 
 | |
5DY4
 
 | | Crystal structure of human Sirt2 in complex with a brominated 2nd generation SirReal inhibitor and NAD+ | | Descriptor: | N-{5-[(7-bromonaphthalen-1-yl)methyl]-1,3-thiazol-2-yl}-2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetamide, NAD-dependent protein deacetylase sirtuin-2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-09-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Aminothiazoles as Potent and Selective Sirt2 Inhibitors: A Structure-Activity Relationship Study.
J.Med.Chem., 59, 2016
|
|
2H59
 
 | | Sir2 H116A-deacetylated p53 peptide-3'-o-acetyl ADP ribose | | Descriptor: | (2S,3S,4R,5S)-2-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-4,5-DIHYDROXYTETRAHYDROFURAN-3-YL ACETATE, ADENOSINE-5-DIPHOSPHORIBOSE, Cellular tumor antigen p53, ... | | Authors: | Hoff, K.G, Avalos, J.L, Sens, K, Wolberger, C. | | Deposit date: | 2006-05-25 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Sirtuin Mechanism from Ternary Complexes Containing NAD(+) and Acetylated Peptide.
Structure, 14, 2006
|
|
5DY5
 
 | | Crystal structure of human Sirt2 in complex with a SirReal probe fragment | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-09-24 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Development of an Affinity Probe for Sirtuin 2.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2H2H
 
 | |
2H4H
 
 | | Sir2 H116Y mutant-p53 peptide-NAD | | Descriptor: | Cellular tumor antigen p53, NAD-dependent deacetylase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hoff, K.G, Avalos, J.L, Sens, K, Wolberger, C. | | Deposit date: | 2006-05-24 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Insights into the Sirtuin Mechanism from Ternary Complexes Containing NAD(+) and Acetylated Peptide.
Structure, 14, 2006
|
|
4IF6
 
 | |
4JT9
 
 | | Crystal Structure of human SIRT3 with ELT inhibitor 3 [14-(4-{2-[(methylsulfonyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide] | | Descriptor: | 4-(4-{2-[(methylsulfonyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide, GLYCEROL, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Dai, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of Thieno[3,2-d]pyrimidine-6-carboxamides as Potent Inhibitors of SIRT1, SIRT2, and SIRT3.
J.Med.Chem., 56, 2013
|
|
4JT8
 
 | | Crystal Structure of human SIRT3 with ELT inhibitor 28 [4-(4-{2-[(2,2-dimethylpropanoyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide[ | | Descriptor: | 4-(4-{2-[(2,2-dimethylpropanoyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide, NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ... | | Authors: | Dai, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of Thieno[3,2-d]pyrimidine-6-carboxamides as Potent Inhibitors of SIRT1, SIRT2, and SIRT3.
J.Med.Chem., 56, 2013
|
|
4HD8
 
 | | Crystal structure of human Sirt3 in complex with Fluor-de-Lys peptide and piceatannol | | Descriptor: | Fluor-de-Lys peptide, ISOPROPYL ALCOHOL, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Steegborn, C. | | Deposit date: | 2012-10-02 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A molecular mechanism for direct sirtuin activation by resveratrol.
Plos One, 7, 2012
|
|
4IG9
 
 | |
4JSR
 
 | | Crystal Structure of human SIRT3 with ELT inhibitor 11c [N-{2-[1-(6-carbamoylthieno[3,2-d]pyrimidin-4-yl)piperidin-4-yl]ethyl}-N'-ethylthiophene-2,5-dicarboxamide] | | Descriptor: | N-{2-[1-(6-carbamoylthieno[3,2-d]pyrimidin-4-yl)piperidin-4-yl]ethyl}-N'-ethylthiophene-2,5-dicarboxamide, NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ... | | Authors: | Dai, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Thieno[3,2-d]pyrimidine-6-carboxamides as Potent Inhibitors of SIRT1, SIRT2, and SIRT3.
J.Med.Chem., 56, 2013
|
|
4HDA
 
 | |
4I5I
 
 | | Crystal structure of the SIRT1 catalytic domain bound to NAD and an EX527 analog | | Descriptor: | (6S)-2-chloro-5,6,7,8,9,10-hexahydrocyclohepta[b]indole-6-carboxamide, NAD-dependent protein deacetylase sirtuin-1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Zhao, X, Allison, D, Condon, B, Zhang, F, Gheyi, T, Zhang, A, Ashok, S, Russell, M, Macewan, I, Qian, Y, Jamison, J.A, Luz, J.G. | | Deposit date: | 2012-11-28 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 angstrom crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition.
J.Med.Chem., 56, 2013
|
|
4L3O
 
 | | Crystal Structure of SIRT2 in complex with the macrocyclic peptide S2iL5 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Yamagata, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-06-06 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Structural Basis for Potent Inhibition of SIRT2 Deacetylase by a Macrocyclic Peptide Inducing Dynamic Structural Change
Structure, 22, 2013
|
|
4KXQ
 
 | | Structure of NAD-dependent protein deacetylase sirtuin-1 (closed state, 1.85 A) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Davenport, A.M, Huber, F.M, Hoelz, A. | | Deposit date: | 2013-05-27 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Structural and Functional Analysis of Human SIRT1.
J.Mol.Biol., 426, 2014
|
|
4BVG
 
 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH NATIVE ALKYLIMIDATE FORMED FROM ACETYL-LYSINE ACS2-PEPTIDE CRYSTALLIZED IN PRESENCE OF THE INHIBITOR EX-527 | | Descriptor: | (2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL ACETATE, 1,2-ETHANEDIOL, ACETYL-COENZYME A SYNTHETASE 2-LIKE, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ex-527 inhibits Sirtuins by exploiting their unique NAD+-dependent deacetylation mechanism.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
4BV2
 
 | | CRYSTAL STRUCTURE OF SIR2 IN COMPLEX WITH THE INHIBITOR EX-527, 2'-O-ACETYL-ADP-RIBOSE AND DEACETYLATED P53-PEPTIDE | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, CELLULAR TUMOR ANTIGEN P53, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4C7B
 
 | | Complex of human Sirt3 with Bromo-Resveratrol and Fluor-De-Lys peptide | | Descriptor: | 5-[(E)-2-(4-bromophenyl)ethenyl]benzene-1,3-diol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-09-20 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Sirt3 Complexes with 4'-Bromo-Resveratrol Reveal Binding Sites and Inhibition Mechanism.
Chem.Biol., 20, 2013
|
|
