7B3D
 
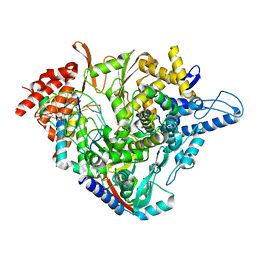 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with AMP at position -4 (structure 3) | | Descriptor: | RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'), RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3'), SARS-CoV-2 RNA-dependent RNA polymerase nsp12, ... | | Authors: | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | Deposit date: | 2020-11-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
8UUH
 
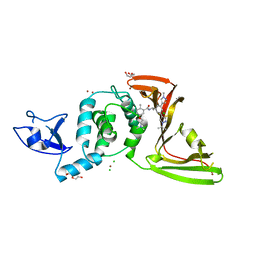 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun12199 | | Descriptor: | 5-[2-(dimethylamino)ethoxy]-2-methyl-N-[(1R)-1-{(3M,5P)-3-(1-methyl-1H-pyrazol-4-yl)-5-[1-(propan-2-yl)-1H-pyrazol-4-yl]phenyl}ethyl]benzamide, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
7BV1
 
 | | Cryo-EM structure of the apo nsp12-nsp7-nsp8 complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
8HOZ
 
 | |
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
8UUF
 
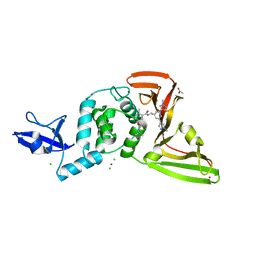 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun11941 | | Descriptor: | ACETATE ION, CHLORIDE ION, N-{(1R)-1-[(3M,5P)-3,5-bis(1-methyl-1H-pyrazol-4-yl)phenyl]ethyl}-5-[2-(dimethylamino)ethoxy]-2-methylbenzamide, ... | | Authors: | Ansari, A, Tan, B, Riuz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
7NT1
 
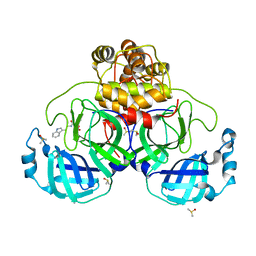 | | Crystal structure of SARS CoV2 main protease in complex with FSP007 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, [(2R)-1-[2-(1H-indol-3-yl)ethylamino]-1-oxidanylidene-butan-2-yl] prop-2-enoate | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6WIQ
 
 | | Crystal structure of the co-factor complex of NSP7 and the C-terminal domain of NSP8 from SARS CoV-2 | | Descriptor: | Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
7CAM
 
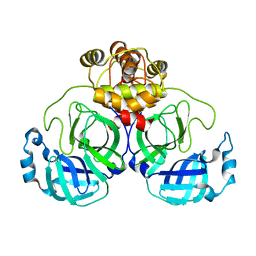 | | SARS-CoV-2 main protease (Mpro) apo structure (space group P212121) | | Descriptor: | 3C-like proteinase | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chou, Y.Z, Chen, Y. | | Deposit date: | 2020-06-09 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.
Am J Cancer Res, 10, 2020
|
|
8UVM
 
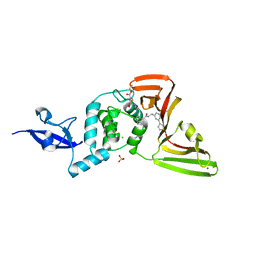 | | SARS-CoV-2 papain-like protease (PLpro) complex with covalent inhibitor Jun11313 | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION, ... | | Authors: | Ansari, A, Tan, B, Chopra, A, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-03 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8DGB
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Q192T Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
7O80
 
 | | Rabbit 80S ribosome in complex with eRF1 and ABCE1 stalled at the STOP codon in the mutated SARS-CoV-2 slippery site | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7LOS
 
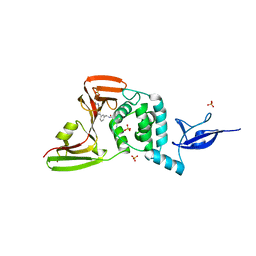 | | SARS-CoV-2 papain-like protease (PLpro) bound to inhibitor XR8-65 | | Descriptor: | 5-(azetidin-3-ylamino)-2-methyl-~{N}-[(1~{R})-1-[3-[5-[[[(3~{R})-oxolan-3-yl]amino]methyl]thiophen-2-yl]phenyl]ethyl]benzamide, CHLORIDE ION, Non-structural protein 3, ... | | Authors: | Ratia, K.M, Xiong, R, Thatcher, G.R. | | Deposit date: | 2021-02-10 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures.
Biorxiv, 2021
|
|
7LLZ
 
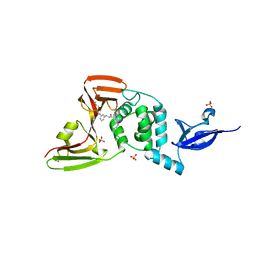 | | SARS-CoV-2 papain-like protease (PLpro) bound to inhibitor XR8-69 | | Descriptor: | N-[(1R)-1-(3-{5-[(acetylamino)methyl]thiophen-2-yl}phenyl)ethyl]-5-[(azetidin-3-yl)amino]-2-methylbenzamide, Non-structural protein 3, SULFATE ION, ... | | Authors: | Ratia, K.M, Xiong, R, Thatcher, G.R. | | Deposit date: | 2021-02-04 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures.
Biorxiv, 2021
|
|
7CXM
 
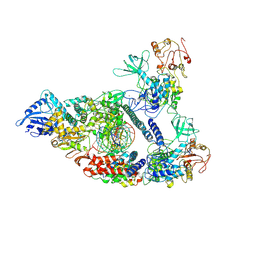 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
6XA9
 
 | | SARS CoV-2 PLpro in complex with ISG15 C-terminal domain propargylamide | | Descriptor: | GLYCEROL, ISG15 CTD-propargylamide, Non-structural protein 3, ... | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
6ZP4
 
 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 2 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
8SQ9
 
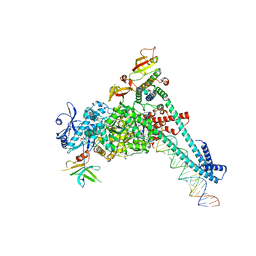 | | SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]uridine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8GW4
 
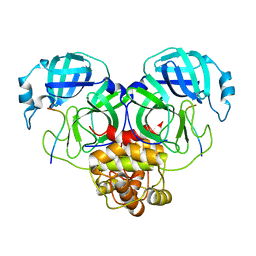 | | SARS-CoV-2 Mpro 1-302/C145A in complex with peptide 8-1 | | Descriptor: | Replicase polyprotein 1ab, peptide 8-1 | | Authors: | Liu, M, Huang, H. | | Deposit date: | 2022-09-16 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The S1'-S3' Pocket of the SARS-CoV-2 Main Protease Is Critical for Substrate Selectivity and Can Be Targeted with Covalent Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8GWS
 
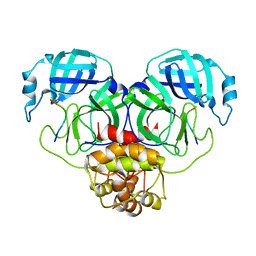 | | SARS-CoV-2 Mpro 1-302 c145a in complex with peptide 4 | | Descriptor: | Replicase polyprotein 1ab, VAL-LYS-LEU-GLN-ALA-ILE-PHE-ARG | | Authors: | Liu, M, Fu, Z, Huang, H. | | Deposit date: | 2022-09-17 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The S1'-S3' Pocket of the SARS-CoV-2 Main Protease Is Critical for Substrate Selectivity and Can Be Targeted with Covalent Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8GWJ
 
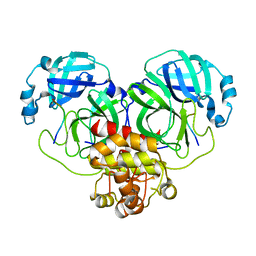 | |
6YYT
 
 | | Structure of replicating SARS-CoV-2 polymerase | | Descriptor: | RNA product, ZINC ION, nsp12, ... | | Authors: | Hillen, H.S, Kokic, G, Farnung, L, Dienemann, C, Tegunov, D, Cramer, P. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of replicating SARS-CoV-2 polymerase.
Nature, 584, 2020
|
|
7K5I
 
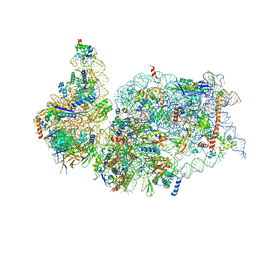 | | SARS-COV-2 nsp1 in complex with human 40S ribosome | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Wang, L, Shi, M, Wu, H. | | Deposit date: | 2020-09-16 | | Release date: | 2020-10-14 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism.
Biorxiv, 2020
|
|
7KEG
 
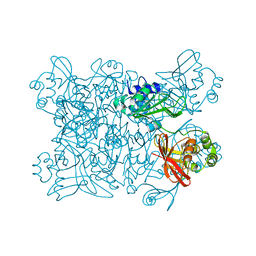 | | Crystal structure from SARS-COV2 NendoU NSP15 | | Descriptor: | PHOSPHATE ION, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | Deposit date: | 2020-10-10 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
6M71
 
 | | SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-01 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
