7ON1
 
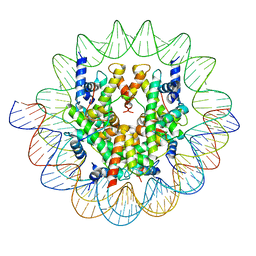 | | Cenp-A nucleosome in complex with Cenp-C | | 分子名称: | BJ4_G0006610.mRNA.1.CDS.1, BJ4_G0007000.mRNA.1.CDS.1, DNA (123-MER), ... | | 著者 | Yan, K, Yang, J, Zhang, Z, Barford, D. | | 登録日 | 2021-05-25 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Cenp-A nucleosome in complex with Cenp-C
To Be Published
|
|
8YTI
 
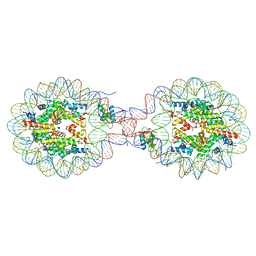 | | Crystal Structure of Nucleosome-H1x Linker Histone Assembly (sticky-169a DNA fragment) | | 分子名称: | CALCIUM ION, CHLORIDE ION, DNA (169-MER), ... | | 著者 | Adhireksan, Z, Qiuye, B, Padavattan, S, Davey, C.A. | | 登録日 | 2024-03-26 | | 公開日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Linker Histones Associate Heterogeneously with Nucleosomes in the Condensed State
To Be Published
|
|
8GRM
 
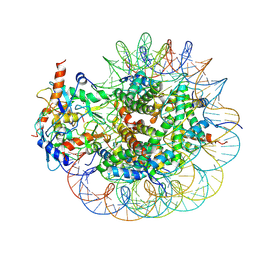 | | Cryo-EM structure of PRC1 bound to H2AK119-UbcH5b-Ub nucleosome | | 分子名称: | COMMD3 protein, DNA (144-MER), DNA (145-MER), ... | | 著者 | Ai, H.S, Zebin, T, Zhihend, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | 登録日 | 2022-09-02 | | 公開日 | 2023-04-12 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
9IPU
 
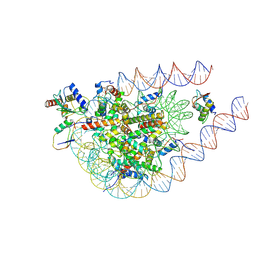 | | cryo-EM structure of the RNF168(1-193)/UbcH5c-Ub ubiquitylation module bound to H1.0-K63-Ub3 modified chromatosome | | 分子名称: | DNA (171-MER), E3 ubiquitin-protein ligase RNF168, Histone H1.0, ... | | 著者 | Ai, H.S, Deng, Z.H, Liu, L. | | 登録日 | 2024-07-11 | | 公開日 | 2024-10-02 | | 最終更新日 | 2025-04-16 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Promotion of RNF168-Mediated Nucleosomal H2A Ubiquitylation by Structurally Defined K63-Polyubiquitylated Linker Histone H1.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8W9C
 
 | |
3G86
 
 | | Hepatitis C virus polymerase NS5B (BK 1-570) with thiazine inhibitor | | 分子名称: | N-{3-[6-fluoro-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl]-1,1-dioxido-4H-1,4-benzothiazin-7-yl}methanesulfonamide, NICKEL (II) ION, RNA-directed RNA polymerase | | 著者 | Harris, S.F, Ghate, M. | | 登録日 | 2009-02-11 | | 公開日 | 2009-09-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 2: Synthesis and structure-activity relationships of benzothiazine-substituted quinolinediones
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6W4S
 
 | | Structure of apo human ferroportin in lipid nanodisc | | 分子名称: | Fab45D8 Heavy Chain, Fab45D8 Light Chain, Solute carrier family 40 member 1 | | 著者 | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | 登録日 | 2020-03-11 | | 公開日 | 2020-09-09 | | 最終更新日 | 2025-05-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
6WBV
 
 | | Structure of human ferroportin bound to hepcidin and cobalt in lipid nanodisc | | 分子名称: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, COBALT (II) ION, Fab45D8 Heavy Chain, ... | | 著者 | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | 登録日 | 2020-03-27 | | 公開日 | 2020-09-09 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
5CIY
 
 | | Structural basis of the recognition of H3K36me3 by DNMT3B PWWP domain | | 分子名称: | DNA (5'-D(P*CP*CP*AP*TP*GP*CP*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*AP*GP*(3DR)P*GP*CP*AP*TP*GP*G)-3'), Modification methylase HhaI, ... | | 著者 | Rondelet, G, DAL MASO, T, Willems, L, Wouters, J. | | 登録日 | 2015-07-13 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.594 Å) | | 主引用文献 | Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B.
J.Struct.Biol., 194, 2016
|
|
3U17
 
 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-(p-benzoyl)phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-(4-benzoylphenyl)-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | 著者 | Gulick, A.M, Drake, E.J, Aldrich, C.C, Neres, J. | | 登録日 | 2011-09-29 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Non-nucleoside inhibitors of BasE, an adenylating enzyme in the siderophore biosynthetic pathway of the opportunistic pathogen Acinetobacter baumannii.
J.Med.Chem., 56, 2013
|
|
3H5S
 
 | | Hepatitis C virus polymerase NS5B with saccharin inhibitor | | 分子名称: | (5S)-5-tert-butyl-1-(4-fluoro-3-methylbenzyl)-4-hydroxy-3-[8-(methylsulfonyl)-1,1-dioxido-6,7,8,9-tetrahydroisothiazolo[4,5-h]isoquinolin-3-yl]-1,5-dihydro-2H-pyrrol-2-one, RNA-directed RNA polymerase | | 著者 | Harris, S.F, Wong, A. | | 登録日 | 2009-04-22 | | 公開日 | 2009-09-08 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 4: structure-based design, synthesis, and biological evaluation of benzo[d]isothiazole-1,1-dioxides
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1YUY
 
 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE GENOTYPE 2a | | 分子名称: | RNA-Dependent RNA polymerase, SULFATE ION | | 著者 | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | 登録日 | 2005-02-14 | | 公開日 | 2005-03-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
3H5U
 
 | | Hepatitis C virus polymerase NS5B with saccharin inhibitor 1 | | 分子名称: | N-({3-[(5S)-5-tert-butyl-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]-1,1-dioxido-1,2-benzisothiazol-7-yl}methyl)methanesulfonamide, RNA-directed RNA polymerase | | 著者 | Harris, S.F, Ghate, M. | | 登録日 | 2009-04-22 | | 公開日 | 2009-09-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 4: structure-based design, synthesis, and biological evaluation of benzo[d]isothiazole-1,1-dioxides
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6KBB
 
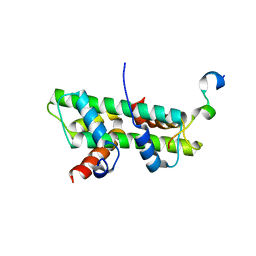 | |
1YV2
 
 | | Hepatitis C virus NS5B RNA-dependent RNA Polymerase genotype 2a | | 分子名称: | GLYCEROL, RNA dependent RNA polymerase, SULFATE ION | | 著者 | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | 登録日 | 2005-02-14 | | 公開日 | 2005-03-22 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structures of the RNA-dependent RNA Polymerase Genotype 2a of Hepatitis C Virus Reveal Two Conformations and Suggest Mechanisms of Inhibition by Non-nucleoside Inhibitors
J.Biol.Chem., 280, 2005
|
|
3U16
 
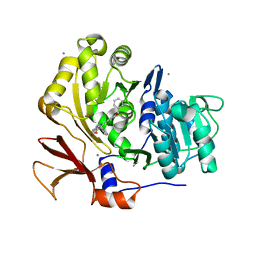 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-(p-benzyloxy)phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid. | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-[4-(benzyloxy)phenyl]-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | 著者 | Gulick, A.M, Drake, E.J, Aldrich, C.C, Neres, J. | | 登録日 | 2011-09-29 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Non-nucleoside inhibitors of BasE, an adenylating enzyme in the siderophore biosynthetic pathway of the opportunistic pathogen Acinetobacter baumannii.
J.Med.Chem., 56, 2013
|
|
5CIU
 
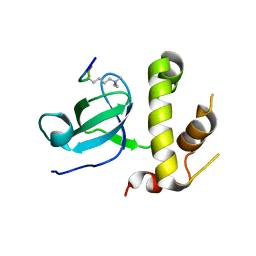 | | Structural basis of the recognition of H3K36me3 by DNMT3B PWWP domain | | 分子名称: | DNA (cytosine-5)-methyltransferase 3B, GLYCEROL, Histone H3.2 | | 著者 | Rondelet, G, DAL MASO, T, Willems, L, Wouters, J. | | 登録日 | 2015-07-13 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B.
J.Struct.Biol., 194, 2016
|
|
2AX1
 
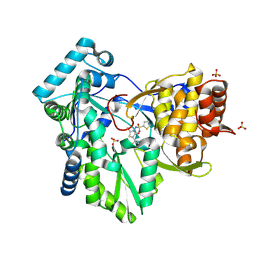 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5ee) | | 分子名称: | 5R-(3,4-DICHLOROPHENYLMETHYL)-3-(2-THIOPHENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | 著者 | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | 登録日 | 2005-09-02 | | 公開日 | 2006-01-24 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
8RHN
 
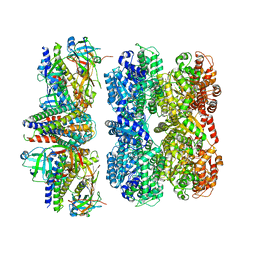 | | Structure of the 55LCC ATPase complex | | 分子名称: | ATPase family gene 2 protein homolog A, ATPase family gene 2 protein homolog B, Cyclin-dependent kinase 2-interacting protein, ... | | 著者 | Foglizzo, M, Degtjarik, O, Zeqiraj, E. | | 登録日 | 2023-12-15 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | The SPATA5-SPATA5L1 ATPase complex directs replisome proteostasis to ensure genome integrity.
Cell, 187, 2024
|
|
4LJY
 
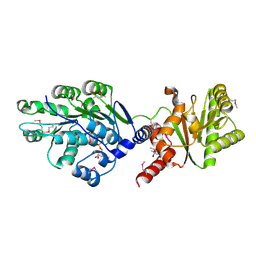 | | Crystal structure of RNA splicing effector Prp5 in complex with ADP | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Zhang, Z.-M, Li, J, Yang, F, Xu, Y, Zhou, J. | | 登録日 | 2013-07-05 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of Prp5p reveals interdomain interactions that impact spliceosome assembly.
Cell Rep, 5, 2013
|
|
4LGE
 
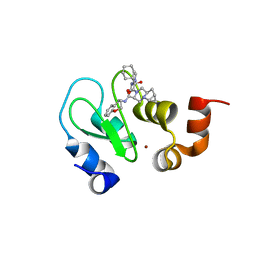 | | Crystal structure of clAP1 BIR3 bound to T3261256 | | 分子名称: | (4S,6aR,7aS)-5-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(4R)-3,4-dihydro-2H-chromen-4-yl]octahydro-1H-cyclopropa[4,5]pyrrolo[1,2-a]pyrazine-4-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | 著者 | Dougan, D.R. | | 登録日 | 2013-06-27 | | 公開日 | 2013-08-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Design, stereoselective synthesis, and biological evaluation of novel tri-cyclic compounds as inhibitor of apoptosis proteins (IAP) antagonists.
Bioorg.Med.Chem., 21, 2013
|
|
4LK2
 
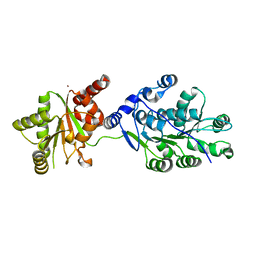 | | Crystal structure of RNA splicing effector Prp5 | | 分子名称: | NICKEL (II) ION, Pre-mRNA-processing ATP-dependent RNA helicase PRP5 | | 著者 | Zhang, Z.-M, Li, J, Yang, F, Xu, Y, Zhou, J. | | 登録日 | 2013-07-05 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Crystal structure of Prp5p reveals interdomain interactions that impact spliceosome assembly.
Cell Rep, 5, 2013
|
|
4LGU
 
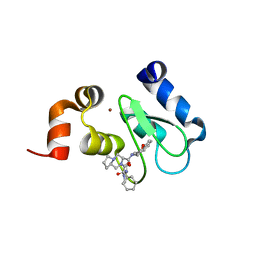 | | Crystal structure of clAP1 BIR3 bound to T3226692 | | 分子名称: | (3S,8aR)-N-((R)-chroman-4-yl)-2-((S)-2-cyclohexyl-2-((S)-2-(methylamino)propanamido)acetyl)octahydropyrrolo[1,2-a]pyrazine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | 著者 | Dougan, D.R, Mol, C.D, Snell, G.P. | | 登録日 | 2013-06-28 | | 公開日 | 2013-08-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design, stereoselective synthesis, and biological evaluation of novel tri-cyclic compounds as inhibitor of apoptosis proteins (IAP) antagonists.
Bioorg.Med.Chem., 21, 2013
|
|
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | 分子名称: | DNA (168-MER), Histone H2A, Histone H2B, ... | | 著者 | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | 登録日 | 2020-08-14 | | 公開日 | 2020-10-21 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
6R87
 
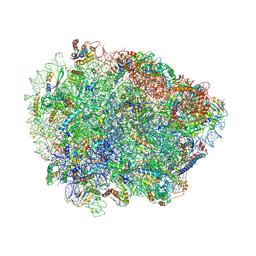 | | Yeast Vms1 (Q295L)-60S ribosomal subunit complex (pre-state without Arb1) | | 分子名称: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Su, T, Izawa, T, Cheng, J, Yamashita, Y, Berninghausen, O, Inada, T, Neupert, W, Beckmann, R. | | 登録日 | 2019-03-31 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis.
Nature, 570, 2019
|
|
