8PHZ
 
 | | Helical reconstruction of CHIKV nsP3 helical scaffolds | | Descriptor: | Non-structural protein 3, ZINC ION | | Authors: | Reguera, J, Hons, M, Zimberger, C, Ptchelkine, D, Jones, R, Desfosses, A. | | Deposit date: | 2023-06-20 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | The alphavirus nsP3 protein forms helical tubular scaffolds important for viral replication and particle assembly
To be published
|
|
8QE8
 
 | | Structure of the non-canonical CTLH E3 substrate receptor WDR26 bound to NMNAT1 substrate | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide/nicotinic acid mononucleotide adenylyltransferase 1, WD repeat-containing protein 26, ... | | Authors: | Chrustowicz, J, Sherpa, D, Schulman, B.A. | | Deposit date: | 2023-08-30 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Non-canonical substrate recognition by the human WDR26-CTLH E3 ligase regulates prodrug metabolism.
Mol.Cell, 84, 2024
|
|
5WNW
 
 | | Chaperone Spy bound to Im7 6-45 ensemble | | Descriptor: | CHLORIDE ION, Colicin-E7 immunity protein, IMIDAZOLE, ... | | Authors: | Horowitz, S, Salmon, L, Koldewey, P, Ahlstrom, L.S, Martin, R, Xu, Q, Afonine, P.V, Trievel, R.C, Brooks, C.L, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
4ZLK
 
 | | Crystal structure of mouse myosin-5a in complex with calcium-bound calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, Unconventional myosin-Va | | Authors: | Shen, M, Zhang, N, Zheng, S, Zhang, W.-B, Zhang, H.-M, Lu, Z, Su, Q.P, Sun, Y, Ye, K, Li, X.-D. | | Deposit date: | 2015-05-01 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural basis for calcium regulation of myosin 5 motor function
To Be Published
|
|
7ZPV
 
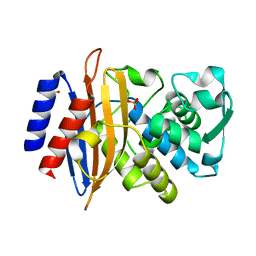 | | Room temperature SSX crystal structure of CTX-M-14 | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Oberthuer, D, Perbandt, M, Prester, A, Rohde, H, Betzel, C, Yefanov, O. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
4ZJV
 
 | |
4U7Z
 
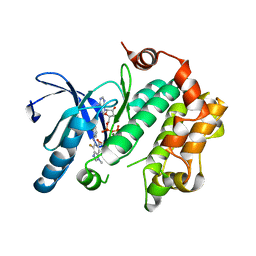 | | Mitogen-Activated Protein Kinase Kinase (MEK1) bound to G805 | | Descriptor: | 5-[(4-bromo-2-chlorophenyl)amino]-4-fluoro-N-(2-hydroxyethoxy)-1-methyl-1H-benzimidazole-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Robarge, K.D, Ultsch, M.H, Wiesmann, C. | | Deposit date: | 2014-07-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Structure based design of novel 6,5 heterobicyclic mitogen-activated protein kinase kinase (MEK) inhibitors leading to the discovery of imidazo[1,5-a] pyrazine G-479.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3UNE
 
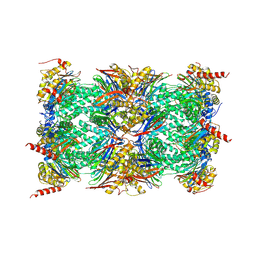 | | Mouse constitutive 20S proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
5WPQ
 
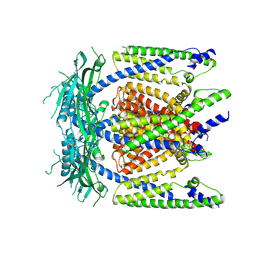 | | Cryo-EM structure of mammalian endolysosomal TRPML1 channel in nanodiscs in closed I conformation at 3.64 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Chen, Q, She, J, Guo, J, Bai, X, Jiang, Y. | | Deposit date: | 2017-08-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.
Nature, 550, 2017
|
|
3UMR
 
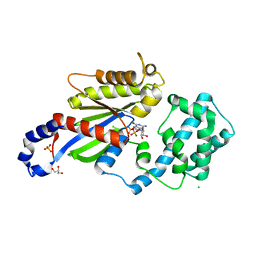 | | Crystal structure of the G202D mutant of human G-alpha-i1 | | Descriptor: | CHLORIDE ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Johnston, C.A, Afshar, K, Snyder, J.T, Tall, G.G, Gonczy, P, Siderovski, D.P, Willard, F.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Determinants Underlying the Temperature-sensitive Nature of a G-alpha Mutant in Asymmetric Cell Division of Caenorhabditis elegans
To be Published
|
|
5X27
 
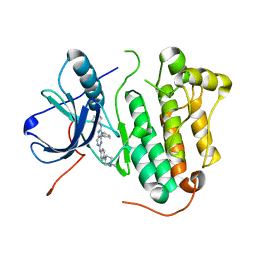 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(5) | | Descriptor: | 9-cyclopentyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
4U80
 
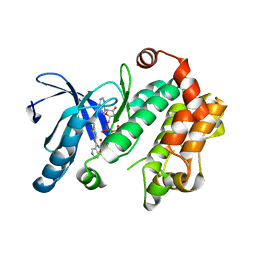 | | MEK 1 kinase bound to G799 | | Descriptor: | 3-[(4-cyclopropyl-2-fluorophenyl)amino]-N-(2-hydroxyethoxy)furo[3,2-c]pyridine-2-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Ultsch, M.H, Robarge, K.D, Weismann, C. | | Deposit date: | 2014-07-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure based design of novel 6,5 heterobicyclic mitogen-activated protein kinase kinase (MEK) inhibitors leading to the discovery of imidazo[1,5-a] pyrazine G-479.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4FID
 
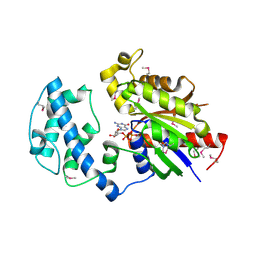 | | Crystal structure of a heterotrimeric G-Protein subunit from entamoeba histolytica, EHG-ALPHA-1 | | Descriptor: | G protein alpha subunit, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Bosch, D.E, Kimple, A.J, Muller, R.E, Gigure, P.M, Willard, F.S, Machius, M, Temple, B.R, Siderovski, D.P. | | Deposit date: | 2012-06-08 | | Release date: | 2012-11-28 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Heterotrimeric G-protein Signaling Is Critical to Pathogenic Processes in Entamoeba histolytica.
Plos Pathog., 8, 2012
|
|
5WTE
 
 | | Cryo-EM structure for Hepatitis A virus full particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6NUS
 
 | |
7UCW
 
 | | Structure of mouse Decr1 in complex with 2'-5' oligoadenylate | | Descriptor: | Decr1 protein, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl] phosphono hydrogen phosphate | | Authors: | Govande, A.A, Kranzusch, P.J. | | Deposit date: | 2022-03-17 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | RNase L-activating 2'-5' oligoadenylates bind ABCF1, ABCF3 and Decr-1.
J.Gen.Virol., 104, 2023
|
|
4GZY
 
 | | Crystal structures of bacterial RNA Polymerase paused elongation complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Weixlbaumer, A, Leon, K, Landick, R, Darst, S.A. | | Deposit date: | 2012-09-06 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5054 Å) | | Cite: | Structural basis of transcriptional pausing in bacteria.
Cell(Cambridge,Mass.), 152, 2013
|
|
4TV1
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with propylparaben | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Estrogen receptor, ... | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2014-06-25 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A structural perspective on nuclear receptors as targets of environmental compounds.
Acta Pharmacol.Sin., 36, 2015
|
|
5Z2T
 
 | | Crystal structure of DNA-bound DUX4-HD2 | | Descriptor: | 5'-D(*TP*TP*CP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*TP*T)-3', 5'-D(P*AP*AP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*GP*T)-3', Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-04 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
5DGG
 
 | | Central domain of uncharacterized Lpg1148 protein from Legionella pneumophila | | Descriptor: | CHLORIDE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Evdokimova, E, Yim, V, Joachimiak, A, Ensminger, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol. Syst. Biol., 12, 2016
|
|
4YKK
 
 | |
4TUZ
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with alpha-zearalenol | | Descriptor: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, 1,2-ETHANEDIOL, Estrogen receptor, ... | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2014-06-25 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural perspective on nuclear receptors as targets of environmental compounds.
Acta Pharmacol. Sin., 36, 2015
|
|
5DJ1
 
 | | Structure of the PLP-Dependent L-Arginine Hydroxylase MppP Holoenzyme | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2015-09-01 | | Release date: | 2015-11-25 | | Last modified: | 2017-05-03 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Streptomyces wadayamensis MppP Is a Pyridoxal 5'-Phosphate-Dependent l-Arginine alpha-Deaminase, gamma-Hydroxylase in the Enduracididine Biosynthetic Pathway.
Biochemistry, 54, 2015
|
|
5DFR
 
 | |
4HB3
 
 | |
