8A1K
 
 | | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with ebsulfur analogue 15 | | Descriptor: | 1,2-ETHANEDIOL, 4,5-bis(chloranyl)-N-(2-hydroxyethyl)-2-sulfanyl-benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | de Munnik, M, Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-throughput screen with the l,d-transpeptidase Ldt Mt2 of Mycobacterium tuberculosis reveals novel classes of covalently reacting inhibitors.
Chem Sci, 14, 2023
|
|
8TQ8
 
 | | Crystal structure of Fab.34.5.8 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab.34.5.8 Heavy chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8A2C
 
 | | The crystal structure of the S178A mutant of PET40, a PETase enzyme from an unclassified Amycolatopsis | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Costanzi, E, Applegate, V, Port, A, Smits, S.H.J. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The metagenome-derived esterase PET40 is highly promiscuous and hydrolyses polyethylene terephthalate (PET).
Febs J., 291, 2024
|
|
115D
 
 | | ORDERED WATER STRUCTURE IN AN A-DNA OCTAMER AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*GP*(BRU)P*AP*(BRU)P*AP*CP*C)-3') | | Authors: | Kennard, O, Cruse, W.B.T, Nachman, J, Prange, T, Shakked, Z, Rabinovich, D. | | Deposit date: | 1993-02-12 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ordered water structure in an A-DNA octamer at 1.7 A resolution.
J.Biomol.Struct.Dyn., 3, 1986
|
|
4RDD
 
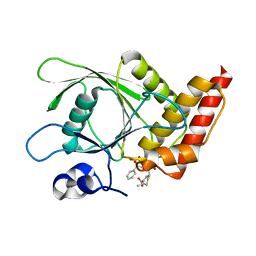 | | Co-crystal structure of SHP2 in complex with a Cefsulodin derivative | | Descriptor: | 1-({(2R)-4-carboxy-2-[(R)-carboxy{[(2R)-2-phenyl-2-sulfoacetyl]amino}methyl]-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyr idinium, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.Y, Yu, Z.H, He, R, Zhang, R.Y. | | Deposit date: | 2014-09-18 | | Release date: | 2015-07-01 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Exploring the Existing Drug Space for Novel pTyr Mimetic and SHP2 Inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
7OV3
 
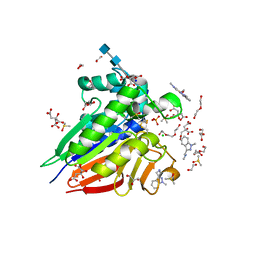 | | Crystal structure of pig purple acid phosphatase in complex with Maybridge fragment CC063346, dimethyl sulfoxide and citrate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, McGeary, R.P, Guddat, L.W, Schenk, G. | | Deposit date: | 2021-06-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational Design of Potent Inhibitors of a Metallohydrolase Using a Fragment-Based Approach.
Chemmedchem, 16, 2021
|
|
7OV2
 
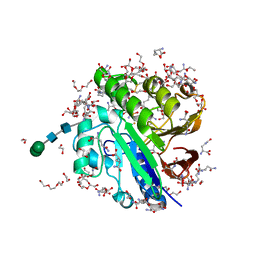 | | Crystal structure of pig purple acid phosphatase in complex with L-glutamine, (poly)ethylene glycol fragments and glycerol | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, FE (III) ION, ... | | Authors: | Feder, D, McGeary, R.P, Guddat, L.W, Schenk, G. | | Deposit date: | 2021-06-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Design of Potent Inhibitors of a Metallohydrolase Using a Fragment-Based Approach.
Chemmedchem, 16, 2021
|
|
7MCF
 
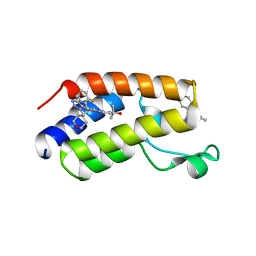 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-6-fluoro-5-[(S)-(3-fluoropyridin-2-yl)(oxan-4-yl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol | | Descriptor: | 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-6-fluoro-5-[(S)-(3-fluoropyridin-2-yl)(oxan-4-yl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol, Bromodomain-containing protein 4 | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-02 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Development of BET inhibitors as potential treatments for cancer: A search for structural diversity.
Bioorg.Med.Chem.Lett., 44, 2021
|
|
8A1J
 
 | | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with maleimide analogue 3 | | Descriptor: | 1,2-ETHANEDIOL, 1-phenylpyrrolidine-2,5-dione, DIMETHYL SULFOXIDE, ... | | Authors: | de Munnik, M, Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | High-throughput screen with the l,d-transpeptidase Ldt Mt2 of Mycobacterium tuberculosis reveals novel classes of covalently reacting inhibitors.
Chem Sci, 14, 2023
|
|
8D1L
 
 | | hBest1 Ca2+-bound partially open aperture state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
6IGI
 
 | | Crystal structure of FT condition 2 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
8RI4
 
 | | Crystal structure of the SARS-CoV-2 Main Protease inhibited by (2-methylsulfanyl-6,7-dihydro-[1,4]dioxino[2,3-f]benzimidazol-3-yl)-(p-tolyl)methanone | | Descriptor: | 3C-like proteinase nsp5, 4-METHYLBENZOIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Charton, J, Deprez, B, Hanoulle, X. | | Deposit date: | 2023-12-18 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the SARS-CoV-2 Main Protease inhibited by (2-methylsulfanyl-6,7-dihydro-[1,4]dioxino[2,3-f]benzimidazol-3-yl)-(p-tolyl)methanone
To Be Published
|
|
6VZ1
 
 | | Cryo-EM structure of human diacylglycerol O-acyltransferase 1 complexed with acyl-CoA substrate | | Descriptor: | Diacylglycerol O-acyltransferase 1, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name), [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Sui, X, Wang, K, Gluchowski, N, Liao, M, Walther, C.T, Farese Jr, V.R. | | Deposit date: | 2020-02-27 | | Release date: | 2020-05-13 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme.
Nature, 581, 2020
|
|
7MS5
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 4-(4-(4-(3,4-difluoro-phenyl)-piperidin-1-ylsulfonyl)-phenyl)-4-oxo-butanoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[4-(3,4-difluorophenyl)piperidine-1-sulfonyl]phenyl}-4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
4R10
 
 | | A conserved phosphorylation switch controls the interaction between cadherin and beta-catenin in vitro and in vivo | | Descriptor: | 1,2-ETHANEDIOL, Cadherin-related hmr-1, Protein humpback-2, ... | | Authors: | Choi, H.-J, Loveless, T, Lynch, A, Bang, I, Hardin, J, Weis, W.I. | | Deposit date: | 2014-08-03 | | Release date: | 2015-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Conserved Phosphorylation Switch Controls the Interaction between Cadherin and beta-Catenin In Vitro and In Vivo
Dev.Cell, 33, 2015
|
|
8AD4
 
 | | X-ray structure of NqrF(129-408)of Vibrio cholerae in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit F, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AD5
 
 | | X-ray structure of NqrF(129-408)of Vibrio cholerae variant F406A | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit F, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6ED6
 
 | |
4R1V
 
 | | Identification and optimization of pyridazinones as potent and selective c-Met kinase inhibitors | | Descriptor: | 3-[1-(3-{5-[(1-methylpiperidin-4-yl)methoxy]pyrimidin-2-yl}benzyl)-6-oxo-1,6-dihydropyridazin-3-yl]benzonitrile, GAMMA-BUTYROLACTONE, Hepatocyte growth factor receptor | | Authors: | Blaukat, A, Bladt, F, Friese-Hamim, M, Knuehl, C, Fittschen, C, Graedler, U, Meyring, M, Dorsch, D, Stieber, F, Schadt, O. | | Deposit date: | 2014-08-07 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Identification and optimization of pyridazinones as potent and selective c-Met kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6Y2J
 
 | | Crystal structure of M. tuberculosis KasA in complex with 4,4,4-trifluoro-N-(isoquinolin-6-yl)butane-1-sulfonamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, 4,4,4-tris(fluoranyl)-~{N}-isoquinolin-6-yl-butane-1-sulfonamide, SODIUM ION, ... | | Authors: | Chung, C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | Exploring the SAR of the beta-Ketoacyl-ACP Synthase Inhibitor GSK3011724A and Optimization around a Genotoxic Metabolite.
Acs Infect Dis., 6, 2020
|
|
8D6F
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with Eph receptor inhibitor / compound 41 | | Descriptor: | (1M)-2-amino-1-(5-hydroxy-2-methylphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, SULFATE ION | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
4R23
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with dicloxacillin | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Dicloxacillin, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-08 | | Release date: | 2014-09-17 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with dicloxacillin
To be Published
|
|
6W3K
 
 | | Structure of unphosphorylated human IRE1 bound to G-9807 | | Descriptor: | 4-[5-[2,6-bis(fluoranyl)phenyl]-2~{H}-pyrazolo[3,4-b]pyridin-3-yl]-2-(4-oxidanylpiperidin-1-yl)-1~{H}-pyrimidin-6-one, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Lammens, A, Wang, W, Ferri, E, Rudolph, J. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
5MZ8
 
 | | Crystal structure of aldehyde dehydrogenase 21 (ALDH21) from Physcomitrella patens in complex with the reaction product succinate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kopecny, D, Vigouroux, A, Briozzo, P, Morera, S. | | Deposit date: | 2017-01-31 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase.
Plant J., 92, 2017
|
|
5KC8
 
 | | Crystal structure of the amino-terminal domain (ATD) of iGluR Delta-2 (GluD2) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glutamate receptor ionotropic, ... | | Authors: | Elegheert, J, Clay, J.E, Siebold, C, Aricescu, A.R. | | Deposit date: | 2016-06-05 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structural basis for integration of GluD receptors within synaptic organizer complexes.
Science, 353, 2016
|
|
