6Z94
 
 | | [4Fe-4S]-dependent thiouracil desulfidase TudS (DUF523Vcz)(S-SAD data) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DUF523 domain-containing protein, ... | | Authors: | Pecqueur, L, Zhou, J, Fontecave, M, Golinelli-Pimpaneau, B. | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Structural Evidence for a [4Fe-5S] Intermediate in the Non-Redox Desulfuration of Thiouracil.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6YWM
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with MES | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Ni, X, Schroeder, M, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6ZEE
 
 | | Structure of PP1(7-300) bound to Phactr1 (507-580) at pH8.4 | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, GLYCEROL, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
4UAI
 
 | | Crystal structure of CXCL12 in complex with inhibitor | | Descriptor: | 1-phenyl-3-[4-(1H-tetrazol-5-yl)phenyl]urea, SULFATE ION, Stromal cell-derived factor 1 | | Authors: | Smith, E.W, Chen, Y. | | Deposit date: | 2014-08-09 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Novel Small Molecule Ligand Bound to the CXCL12 Chemokine.
J.Med.Chem., 57, 2014
|
|
7NIV
 
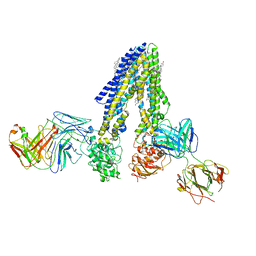 | | Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab and QA2-Fab (phosphatidylcholine-bound, occluded conformation) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4B1 Fab-fragment light chain, CHOLESTEROL, ... | | Authors: | Nosol, K, Locher, K.P. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of ABCB4 provide insight into phosphatidylcholine translocation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NIW
 
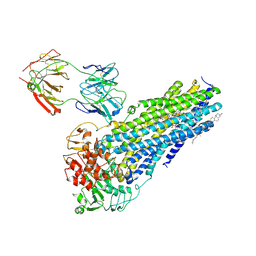 | | Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab (posaconazole-bound, inward-open conformation) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4B1 Fab-fragment heavy chain, 4B1 Fab-fragment light chain, ... | | Authors: | Nosol, K, Locher, K.P. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of ABCB4 provide insight into phosphatidylcholine translocation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Z1H
 
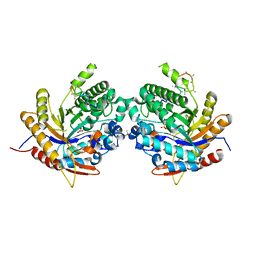 | | Ancestral glycosidase (family 1) | | Descriptor: | ANCESTRAL RECONSTRUCTED GLYCOSIDASE, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Hoshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | Deposit date: | 2020-05-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
7QGP
 
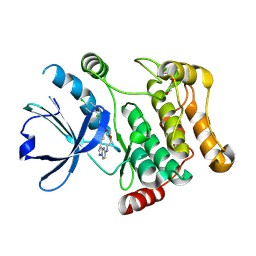 | | STK10 (LOK) bound to Macrocycle CKJB51 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-chloranyl-2-methyl-phenyl)methyl]-3-(7,10-dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.1^{2,6}.0^{17,20}]docosa-1(20),2(22),3,5,14(21),15,18-heptaen-5-yl)urea, CHLORIDE ION, ... | | Authors: | Berger, B.-T, Schroeder, M, Kraemer, A, Schwalm, M.P, Kurz, C.G, Amrhein, J.A, Hanke, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STK10 bound to CKJB51
To Be Published
|
|
6Z4Y
 
 | | Crystal structure of Aurora A (STK6) in complex with macrocycle ODS2003208 | | Descriptor: | 1,2-ETHANEDIOL, 6-(2-methoxyethoxy)-11-methyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, Aurora kinase A, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of Aurora A (STK6) in complex with macrocycle ODS2003208
To Be Published
|
|
6PJ3
 
 | | Crystal structure of the Klebsiella pneumoniae LpxH/JH-LPH-33 complex | | Descriptor: | 1,2-ETHANEDIOL, 1-[5-({4-[3-chloro-5-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7A2H
 
 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6,7-tribromo-1H-imidazo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7-tris(bromanyl)-1~{H}-imidazo[4,5-b]pyridine, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-18 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
8ULG
 
 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
4TYV
 
 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
8YTU
 
 | | Mipa-PETase from Micromonospora pattaloongensis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, cutinase | | Authors: | Hong, H, Seo, H, Park, J, Kim, K.-J. | | Deposit date: | 2024-03-26 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Landscape profiling of PET depolymerases using a natural sequence cluster framework.
Science, 387, 2025
|
|
6XXM
 
 | | Crystal Structure of Human Deoxyhypusine Synthase in complex with putrescine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,4-DIAMINOBUTANE, Deoxyhypusine synthase | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Half Way to Hypusine-Structural Basis for Substrate Recognition by Human Deoxyhypusine Synthase.
Biomolecules, 10, 2020
|
|
6XXJ
 
 | | Crystal Structure of Human Deoxyhypusine Synthase in complex with spermidine and NAD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Half Way to Hypusine-Structural Basis for Substrate Recognition by Human Deoxyhypusine Synthase.
Biomolecules, 10, 2020
|
|
8P0S
 
 | | Crystal structure HR1 domain of Rho-associated coiled-coil protein kinases (ROCK-HR1) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Rho-associated protein kinase 1 | | Authors: | Dubey, B.N, Dvorsky, R, Gremer, L, Vetter, I.R, Schmitt, L, Groth, G, Ahmadian, M.R. | | Deposit date: | 2023-05-10 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the p160 Rho-associated coiled-coil-containing protein kinase
To Be Published
|
|
8OUU
 
 | | Crystal structure of D1228V c-MET bound by compound 29 | | Descriptor: | 1,2-ETHANEDIOL, 5-(3-ethynyl-5-fluoranyl-1H-indazol-7-yl)-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, FORMIC ACID, ... | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OVZ
 
 | | Crystal structure of D1228V c-MET bound by compound 16 | | Descriptor: | 1-[(1S)-1-[3-(1H-imidazol-4-yl)phenyl]ethyl]-5-(1H-indazol-7-yl)pyrimidine-2,4-dione, Hepatocyte growth factor receptor, IODIDE ION | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-26 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OU8
 
 | | Crystal structure of E. coli threonyl tRNA synthetase in complex with a TM84 analogue | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Threonine--tRNA ligase, ... | | Authors: | Rodriguez Buitrago, J.A, Parisini, E, Aigars, J. | | Deposit date: | 2023-04-22 | | Release date: | 2023-07-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis and evaluation of an agrocin 84 toxic moiety (TM84) analogue as a malarial threonyl tRNA synthetase inhibitor.
Org.Biomol.Chem., 21, 2023
|
|
8GY5
 
 | |
6X1B
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with the Product Nucleotide GpU. | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-R(*GP*U)-3'), PHOSPHATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-18 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
4UCP
 
 | |
4XUF
 
 | | Crystal structure of the FLT3 kinase domain bound to the inhibitor quizartinib (AC220) | | Descriptor: | 1-(5-tert-butyl-1,2-oxazol-3-yl)-3-(4-{7-[2-(morpholin-4-yl)ethoxy]imidazo[2,1-b][1,3]benzothiazol-2-yl}phenyl)urea, Receptor-type tyrosine-protein kinase FLT3 | | Authors: | Zorn, J.A, Wang, Q, Fujimura, E, Barros, T, Kuriyan, J. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the FLT3 Kinase Domain Bound to the Inhibitor Quizartinib (AC220).
Plos One, 10, 2015
|
|
1FS3
 
 | |
