4NLV
 
 | | Poliovirus Polymerase - G289A/C290F Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4NRT
 
 | | Human Norovirus polymerase bound to Compound 6 (suramin derivative) | | Descriptor: | 4-({4-methyl-3-[(3-nitrobenzoyl)amino]benzoyl}amino)naphthalene-1,5-disulfonic acid, hNV-RdRp | | Authors: | Croci, R, Pezzullo, M, Tarantino, D, Mastrangelo, E, Milani, M, Bolognesi, M. | | Deposit date: | 2013-11-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | Structural bases of norovirus RNA dependent RNA polymerase inhibition by novel suramin-related compounds.
Plos One, 9, 2014
|
|
4NLW
 
 | | Poliovirus Polymerase - G289A/C290I Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4NLY
 
 | | Poliovirus Polymerase - C290E Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4NLR
 
 | | Poliovirus Polymerase - C290S Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4NLU
 
 | | Poliovirus Polymerase - G289A Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
9UHT
 
 | | SARS-CoV-2 E-RTC in complex with RNA-nsp9 and GMPPNP | | Descriptor: | Helicase nsp13, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Huang, Y.C, Liu, Y.X, Lou, Z.Y, Rao, Z.H, Yan, L.M. | | Deposit date: | 2025-04-14 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Response to Matter Arising: Substrate selection and transition state support the mechanism for GTP-mediate RNA capping catalyzed by SARS-CoV-2 NiRAN
To Be Published
|
|
6YYT
 
 | | Structure of replicating SARS-CoV-2 polymerase | | Descriptor: | RNA product, ZINC ION, nsp12, ... | | Authors: | Hillen, H.S, Kokic, G, Farnung, L, Dienemann, C, Tegunov, D, Cramer, P. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of replicating SARS-CoV-2 polymerase.
Nature, 584, 2020
|
|
7THM
 
 | | SARS-CoV-2 nsp12/7/8 complex with a native N-terminus nsp9 | | Descriptor: | MANGANESE (II) ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Osinski, A, Tagliabracci, V.S, Chen, Z, Li, Y. | | Deposit date: | 2022-01-11 | | Release date: | 2022-03-16 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The mechanism of RNA capping by SARS-CoV-2.
Nature, 609, 2022
|
|
6XQB
 
 | | SARS-CoV-2 RdRp/RNA complex | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-07-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of SARS-CoV-2 RdRp/RNA complex at 3.4 Angstroms resolution
To Be Published
|
|
6XEZ
 
 | | Structure of SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Llewellyn, E.C, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex.
Cell, 182, 2020
|
|
8XCH
 
 | |
7B3D
 
 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with AMP at position -4 (structure 3) | | Descriptor: | RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'), RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3'), SARS-CoV-2 RNA-dependent RNA polymerase nsp12, ... | | Authors: | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | Deposit date: | 2020-11-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
7B3B
 
 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with Remdesivir at position -3 (structure 1) | | Descriptor: | DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*G)-D(P*(RMP))-R(P*UP*G)-3'), Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | Deposit date: | 2020-11-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
7B3C
 
 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with Remdesivir at position -4 (structure 2) | | Descriptor: | DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*A)-D(P*(RMP))-R(P*GP*UP*G)-3'), Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | Deposit date: | 2020-11-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
7AAP
 
 | | Nsp7-Nsp8-Nsp12 SARS-CoV2 RNA-dependent RNA polymerase in complex with template:primer dsRNA and favipiravir-RTP | | Descriptor: | MAGNESIUM ION, Non-structural protein 12, Non-structural protein 7, ... | | Authors: | Naydenova, K, Muir, K.W, Wu, L.F, Zhang, Z, Coscia, F, Peet, M, Castro-Hartman, P, Qian, P, Sader, K, Dent, K, Kimanius, D, Sutherland, J.D, Lowe, J, Barford, D, Russo, C.J. | | Deposit date: | 2020-09-04 | | Release date: | 2020-09-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KRN
 
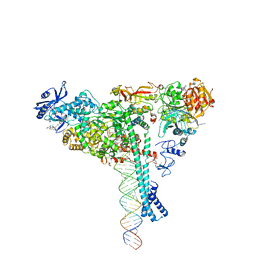 | | Structure of SARS-CoV-2 backtracked complex bound to nsp13 helicase - nsp13(1)-BTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KRP
 
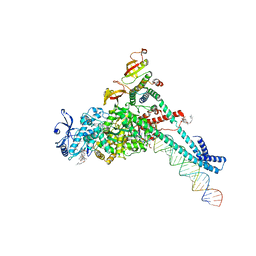 | | Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - BTC (local refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHAPSO, MAGNESIUM ION, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KRO
 
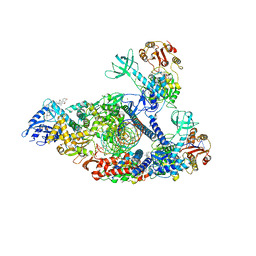 | | Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - nsp13(2)-BTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7L1F
 
 | | SARS-CoV-2 RdRp in complex with 4 Remdesivir monophosphate | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (5'-R(P*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*UP*CP*UP*UP*AP*G)-3'), ... | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2020-12-14 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication.
Mol.Cell, 81, 2021
|
|
9BI9
 
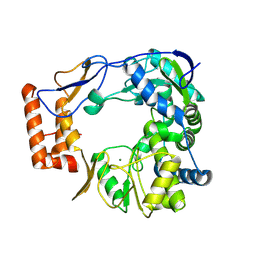 | | GII.4 Sydney 2012 Polymerase domain of ProPol precursor | | Descriptor: | MAGNESIUM ION, RNA-dependent RNA polymerase domain of Protease-Polymerase precursor | | Authors: | Eruera, A, Krause, K. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Activity and cryo-EM structure of the polymerase domain of the human norovirus ProPol precursor.
J.Virol., 98, 2024
|
|
9CPO
 
 | |
9CGV
 
 | | SARS-CoV-2 nsp12 NiRAN domain bound to a covalent inhibitor SW090466-1 | | Descriptor: | MANGANESE (II) ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Osinski, A, Hernandez, G, Tagliabracci, V.S. | | Deposit date: | 2024-07-01 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Covalent inhibition of the SARS-CoV-2 NiRAN domain via an active-site cysteine.
J.Biol.Chem., 301, 2025
|
|
8SQJ
 
 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode | | Descriptor: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8SQ9
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]uridine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
