6RBI
 
 | | Crystal structure of KDM5B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol | | Descriptor: | 1,2-ETHANEDIOL, 5-(1~{H}-1,2,3,4-tetrazol-5-yl)quinolin-8-ol, Lysine-specific demethylase 5B,Lysine-specific demethylase 5B, ... | | Authors: | Johansson, C, Newman, J.A, Kawamura, A, Schofield, C.J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-04-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of KDM5B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol
To Be Published
|
|
2RNY
 
 | | Complex Structures of CBP Bromodomain with H4 ack20 Peptide | | Descriptor: | CREB-binding protein, Histone H4 | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
5HWZ
 
 | | Crystal structure of nitrophorin 4 D30N mutant with nitrite | | Descriptor: | NITRITE ION, Nitrophorin-4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ogata, H, He, C, Lubitz, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Elucidation of the heme active site electronic structure affecting the unprecedented nitrite dismutase activity of the ferrihemebproteins, the nitrophorins.
Chem Sci, 7, 2016
|
|
6RCF
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-15]-OH | | Descriptor: | 2-[(3~{a}~{R},6~{R},8~{a}~{S})-1-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-6-ethyl-8-oxidanylidene-3,3~{a},6,8~{a}-tetrahydro-2~{H}-pyrrolo[2,3-c]azepin-7-yl]ethanoic acid, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5HWO
 
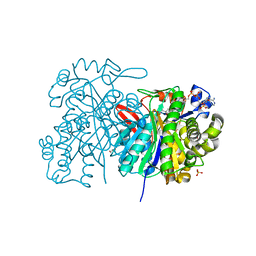 | | MvaS in complex with 3-hydroxy-3-methylglutaryl coenzyme A | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, GLYCEROL, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Bock, T, Kasten, J, Blankenfeldt, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of the HMG-CoA Synthase MvaS from the Gram-Negative Bacterium Myxococcus xanthus.
Chembiochem, 17, 2016
|
|
3QUM
 
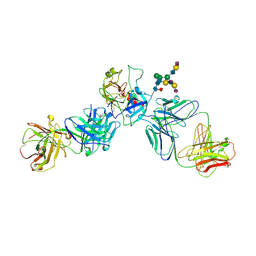 | | Crystal structure of human prostate specific antigen (PSA) in Fab sandwich with a high affinity and a PCa selective antibody | | Descriptor: | Fab 5D3D11 Heavy Chain, Fab 5D3D11 Light Chain, Fab 5D5A5 Heavy Chain, ... | | Authors: | Stura, E.A, Muller, B.H, Michel, S, Ducancel, F. | | Deposit date: | 2011-02-24 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human prostate-specific antigen in a sandwich antibody complex.
J.Mol.Biol., 414, 2011
|
|
8PYI
 
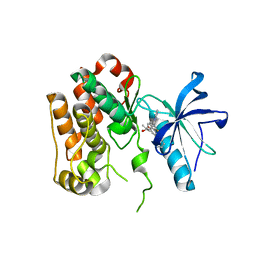 | | Human IGF1R with inhibitor 6 | | Descriptor: | 3-[8-azanyl-1-(4-ethoxy-8-fluoranyl-2-phenyl-quinolin-7-yl)imidazo[1,5-a]pyrazin-3-yl]-1-methyl-cyclobutan-1-ol, Insulin-like growth factor 1 receptor beta chain | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Human IGF1R with inhibitor 6
To Be Published
|
|
6RCJ
 
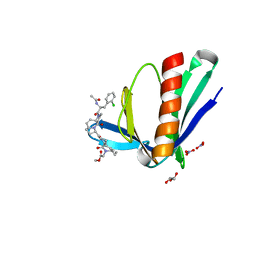 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-15]-OMe | | Descriptor: | GLYCEROL, NITRATE ION, Protein enabled homolog, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5HQI
 
 | | Insulin with proline analog HzP at position B28 in the T2 state | | Descriptor: | Insulin A-Chain, Insulin B-Chain | | Authors: | Lieblich, S.A, Fang, K.Y, Cahn, J.K.B, Tirrell, D.A. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.
J. Am. Chem. Soc., 139, 2017
|
|
6R83
 
 | | CryoEM structure and molecular model of squid hemocyanin (Todarodes pacificus , TpH) | | Descriptor: | Hemocyanin subunit 1 | | Authors: | Tanaka, Y, Kato, S, Stabrin, M, Raunser, S, Matsui, T, Gatsogiannis, C. | | Deposit date: | 2019-03-31 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Cryo-EM reveals the asymmetric assembly of squid hemocyanin.
Iucrj, 6, 2019
|
|
2RQ6
 
 | | Solution structure of the epsilon subunit of the F1-atpase from thermosynechococcus elongatus BP-1 | | Descriptor: | ATP synthase epsilon chain | | Authors: | Yagi, H, Konno, H, Murakami-Fuse, T, Oroguchi, H, Akutsu, T, Ikeguchi, M, Hisabori, T. | | Deposit date: | 2009-03-03 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional analysis of the intrinsic inhibitor subunit epsilon of F1-ATPase from photosynthetic organisms.
Biochem.J., 425, 2010
|
|
5HVI
 
 | |
2SBT
 
 | | A COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF SUBTILISIN BPN AND SUBTILISIN NOVO | | Descriptor: | ACETONE, SUBTILISIN NOVO | | Authors: | Drenth, J, Hol, W.G.J, Jansonius, J.N, Koekoek, R. | | Deposit date: | 1976-09-07 | | Release date: | 1976-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A comparison of the three-dimensional structures of subtilisin BPN' and subtilisin novo.
Cold Spring Harbor Symp.Quant.Biol., 36, 1972
|
|
2ST1
 
 | |
5HW1
 
 | |
8PPL
 
 | | MERS-CoV Nsp1 bound to the human 43S pre-initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
3RUT
 
 | | FXR with SRC1 and GSK359 | | Descriptor: | 6-(4-{[3-(2,6-dichlorophenyl)-5-(propan-2-yl)-1,2-oxazol-4-yl]methoxy}phenyl)-1-benzothiophene-3-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Williams, S.P, Madauss, K.P. | | Deposit date: | 2011-05-05 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformationally constrained farnesoid X receptor (FXR) agonists: Alternative replacements of the stilbene.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RV2
 
 | |
6R3U
 
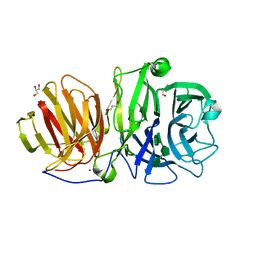 | | Endo-levanase BT1760 mutant E221A from Bacteroides thetaiotaomicron complexed with levantetraose | | Descriptor: | GLYCEROL, Glycoside hydrolase family 32, ZINC ION, ... | | Authors: | Eek, P, Ernits, K, Lukk, T, Alamae, T. | | Deposit date: | 2019-03-21 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First crystal structure of an endo-levanase - the BT1760 from a human gut commensal Bacteroides thetaiotaomicron.
Sci Rep, 9, 2019
|
|
2SNI
 
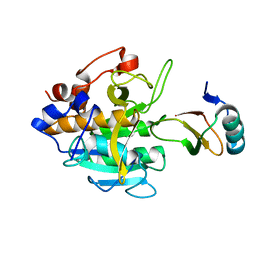 | |
3S1K
 
 | |
2SXL
 
 | | SEX-LETHAL RBD1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SEX-LETHAL PROTEIN | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Kigawa, T, Takio, K, Shimura, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-07-16 | | Release date: | 1998-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A characteristic arrangement of aromatic amino acid residues in the solution structure of the amino-terminal RNA-binding domain of Drosophila sex-lethal.
J.Mol.Biol., 272, 1997
|
|
5HZX
 
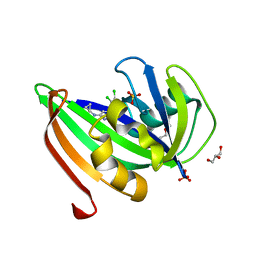 | | Crystal structure of zebrafish MTH1 in complex with TH588 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Narwal, M, Gustafsson, R, Brautigam, L, Pudelko, L, Jemth, A.-S, Gad, H, Karsten, S, Carreras-Puigvert, J, Homan, E, Berndt, C, Berglund, U.W, Helleday, T, Stenmark, P. | | Deposit date: | 2016-02-03 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hypoxic Signaling and the Cellular Redox Tumor Environment Determine Sensitivity to MTH1 Inhibition.
Cancer Res., 76, 2016
|
|
6RAZ
 
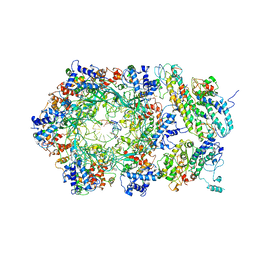 | | D. melanogaster CMG-DNA, State 2B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AT18545p, ... | | Authors: | Eickhoff, P, Martino, F, Costa, A. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-18 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (4.46 Å) | | Cite: | Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome.
Cell Rep, 28, 2019
|
|
6R7N
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-29 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
