4CI3
 
 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to Pomalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Pomalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4CI1
 
 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to thalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Thalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4F52
 
 | | Structure of a Glomulin-RBX1-CUL1 complex | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase RBX1, Glomulin, ... | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2012-05-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a Glomulin-RBX1-CUL1 Complex: Inhibition of a RING E3 Ligase through Masking of Its E2-Binding Surface.
Mol.Cell, 47, 2012
|
|
7OO3
 
 | | Pol II-CSB-CSA-DDB1-UVSSA (Structure1) | | Descriptor: | CSB element, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-26 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OOP
 
 | | Pol II-CSB-CSA-DDB1-UVSSA-PAF-SPT6 (Structure 3) | | Descriptor: | DNA damage-binding protein 1, DNA excision repair protein ERCC-6, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OOB
 
 | | Pol II-CSB-CSA-DDB1-UVSSA-ADPBeF3 (Structure2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA damage-binding protein 1, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
3EI4
 
 | | Structure of the hsDDB1-hsDDB2 complex | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2 | | Authors: | Scrima, A, Pavletich, N.P, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
3EI3
 
 | | Structure of the hsDDB1-drDDB2 complex | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, TETRAETHYLENE GLYCOL | | Authors: | Scrima, A, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
6ZX9
 
 | | Crystal structure of SIV Vpr,fused to T4 lysozyme, isolated from moustached monkey, bound to human DDB1 and human DCAF1 (amino acid residues 1046-1396) | | Descriptor: | DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, GLYCEROL, ... | | Authors: | Schwefel, D, Banchenko, S. | | Deposit date: | 2020-07-29 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.519729 Å) | | Cite: | Structural insights into Cullin4-RING ubiquitin ligase remodelling by Vpr from simian immunodeficiency viruses.
Plos Pathog., 17, 2021
|
|
6ZUE
 
 | |
7PLO
 
 | | H. sapiens replisome-CUL2/LRR1 complex | | Descriptor: | Cell division control protein 45 homolog, Claspin, Cullin-2, ... | | Authors: | Jones, M.J, Yeeles, J.T.P, Deegan, T.D, Jenkyn-Bedford, M. | | Deposit date: | 2021-09-01 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A conserved mechanism for regulating replisome disassembly in eukaryotes.
Nature, 600, 2021
|
|
7B5N
 
 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: NEDD8-CUL1-RBX1-UBE2L3~Ub~ARIH1. | | Descriptor: | 5-azanylpentan-2-one, Cullin-1, E3 ubiquitin-protein ligase ARIH1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
7B5R
 
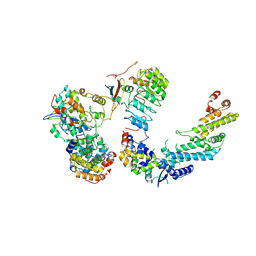 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-SKP1-SKP2-CKSHS1-Cyclin A-CDK2-p27 | | Descriptor: | Cullin-1, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-07 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
7B5S
 
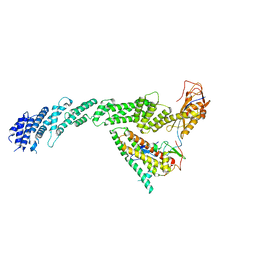 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-ARIH1 Ariadne. Transition State 1 | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase ARIH1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-07 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
7B5L
 
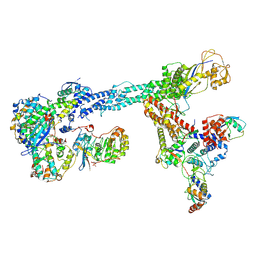 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: NEDD8-CUL1-RBX1-SKP1-SKP2-CKSHS1-Cyclin A-CDK2-p27-UBE2L3~Ub~ARIH1. Transition State 1 | | Descriptor: | 5-azanylpentan-2-one, Cullin-1, Cyclin-A2, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
3DQV
 
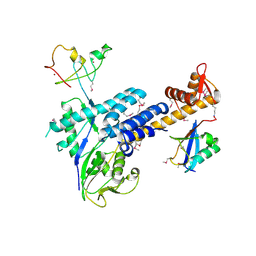 | | Structural Insights into NEDD8 Activation of Cullin-RING Ligases: Conformational Control of Conjugation | | Descriptor: | Cullin-5, NEDD8, Rbx1, ... | | Authors: | Duda, D.M, Borg, L.A, Scott, D.C, Hunt, H.W, Hammel, M, Schulman, B.A. | | Deposit date: | 2008-07-09 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation.
Cell(Cambridge,Mass.), 134, 2008
|
|
7B5M
 
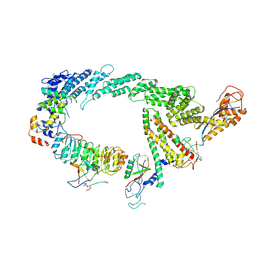 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-SKP1-SKP2-CKSHS1-p27~Ub~ARIH1. Transition State 2 | | Descriptor: | Cullin-1, Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-17 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
3EI1
 
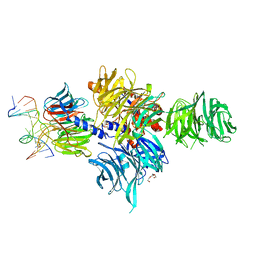 | |
3E0C
 
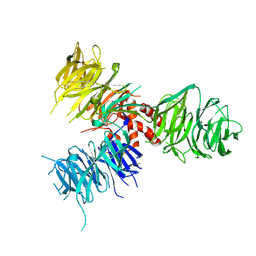 | | Crystal Structure of DNA Damage-Binding protein 1(DDB1) | | Descriptor: | DNA damage-binding protein 1 | | Authors: | Amaya, M.F, Xu, L, Hao, H, Bountra, C, Wickstroem, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
3DPL
 
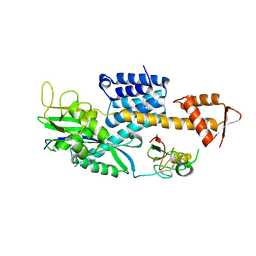 | |
3EI2
 
 | | Structure of hsDDB1-drDDB2 bound to a 16 bp abasic site containing DNA-duplex | | Descriptor: | 5'-D(*DAP*DAP*DAP*DTP*DGP*DAP*DAP*DTP*(3DR)P*DAP*DAP*DGP*DCP*DAP*DGP*DG)-3', 5'-D(*DCP*DCP*DTP*DGP*DCP*DTP*DTP*DTP*DAP*DTP*DTP*DCP*DAP*DTP*DTP*DT)-3', DNA damage-binding protein 1, ... | | Authors: | Scrima, A, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
8PQL
 
 | | K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide | | Descriptor: | 5-azanylpentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-07-11 | | Release date: | 2024-02-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q7R
 
 | | Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide | | Descriptor: | 5-azanyl-1-oxidanyl-pentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
8GQ6
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1 dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Sun, L, Chen, Z, Hu, Y, Mao, Q. | | Deposit date: | 2022-08-29 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QU8
 
 | | PROTAC-mediated complex of KRAS with VHL/Elongin-B/Elongin-C/Cullin-2/Rbx1 | | Descriptor: | (2S,4R)-1-[(2S)-2-[4-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2,3-triazol-1-yl]-3-methyl-butanoyl]-N-[(1R)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-2-oxidanyl-ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Fischer, G, Peter, D, Arce-Solano, S. | | Deposit date: | 2023-10-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
