3N1H
 
 | | Crystal Structure of StWhy2 | | Descriptor: | PHOSPHATE ION, StWhy2 | | Authors: | Cappadocia, L, Brisson, N, Sygusch, J. | | Deposit date: | 2010-05-15 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Crystal Structures of DNA-Whirly Complexes and Their Role in Arabidopsis Organelle Genome Repair.
Plant Cell, 22, 2010
|
|
5HRO
 
 | |
7E1B
 
 | | Crystal structure of VbrR-DNA complex | | Descriptor: | DNA (26-MER), DNA-binding response regulator | | Authors: | Hong, S, Zhang, X, Zhang, P. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (4.587 Å) | | Cite: | Structural basis of phosphorylation-induced activation of the response regulator VbrR.
Acta Biochim.Biophys.Sin., 2023
|
|
6IDO
 
 | | Crystal structure of Klebsiella pneumoniae sigma4 of sigmaS fusing with the RNA polymerase beta-flap-tip-helix in complex with -35 element DNA | | Descriptor: | DNA (5'-D(P*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*AP*AP*TP*CP*G)-3'), DNA (5'-D(P*GP*AP*TP*TP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*C)-3'), RNA polymerase sigma factor RpoS,RNA polymerase beta-flap-tip-helix | | Authors: | Lou, Y.C, Chien, C.Y, Chen, C, Hsu, C.H. | | Deposit date: | 2018-09-10 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.748 Å) | | Cite: | Structural basis for -35 element recognition by sigma4chimera proteins and their interactions with PmrA response regulator.
Proteins, 88, 2020
|
|
5D8K
 
 | |
4HW0
 
 | | Crystal structure of Sso10a-2, a DNA-binding protein from Sulfolobus solfataricus | | Descriptor: | DNA-binding protein Sso10a-2 | | Authors: | Waterreus, W.J, Goosen, N, Moolenaar, G.F, Driessen, R.P.C, Dame, R.T, Pannu, N.S. | | Deposit date: | 2012-11-07 | | Release date: | 2013-10-30 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse architectural properties of Sso10a proteins: Evidence for a role in chromatin compaction and organization.
Sci Rep, 6, 2016
|
|
6JHE
 
 | | Crystal Structure of Bacillus subtilis SigW domain 4 in complexed with -35 element DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*GP*TP*TP*TP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*AP*AP*AP*CP*CP*TP*TP*T)-3'), ECF RNA polymerase sigma factor SigW | | Authors: | Kwon, E, Devkota, S.R, Pathak, D, Dahal, P, Kim, D.Y. | | Deposit date: | 2019-02-18 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Structural analysis of the recognition of the -35 promoter element by SigW from Bacillus subtilis.
Plos One, 14, 2019
|
|
3C58
 
 | | Crystal structure of a complex between the wild-type lactococcus lactis Fpg (MutM) and a N7-Benzyl-Fapy-dG containing DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DCP*DTP*DTP*DTP*(SOS)P*DTP*DTP*DTP*DCP*DTP*DCP*DG)-3'), DNA (5'-D(*DGP*DCP*DGP*DAP*DGP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DGP*DA)-3'), DNA glycosylase, ... | | Authors: | Coste, F, Castaing, B, Carell, T. | | Deposit date: | 2008-01-31 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacterial base excision repair enzyme Fpg recognizes bulky N7-substituted-FapydG lesion via unproductive binding mode
Chem.Biol., 15, 2008
|
|
3V79
 
 | | Structure of human Notch1 transcription complex including CSL, RAM, ANK, and MAML-1 on HES-1 promoter DNA sequence | | Descriptor: | DNA 5'-D(*GP*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*AP*AP*A)-3', DNA 5'-D(*TP*TP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*TP*AP*AP*C)-3', Mastermind-like protein 1, ... | | Authors: | Nam, Y, Sliz, P, Blacklow, S. | | Deposit date: | 2011-12-20 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Conformational Locking upon Cooperative Assembly of Notch Transcription Complexes.
Structure, 20, 2012
|
|
6JBX
 
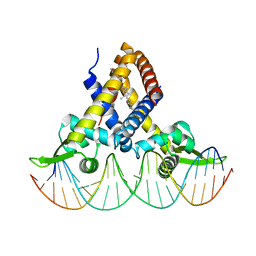 | | Crystal structure of Streptococcus pneumoniae FabT in complex with DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*GP*TP*TP*TP*GP*AP*CP*TP*GP*TP*CP*AP*AP*AP*TP*TP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*AP*AP*TP*TP*TP*GP*AP*CP*AP*GP*TP*CP*AP*AP*AP*CP*TP*AP*TP*T)-3'), Fatty acid biosynthesis transcriptional regulator, ... | | Authors: | Zuo, G, Chen, Z.P, Li, Q, Zhou, C.Z. | | Deposit date: | 2019-01-27 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into repression of the Pneumococcal fatty acid synthesis pathway by repressor FabT and co-repressor acyl-ACP.
Febs Lett., 593, 2019
|
|
8IFO
 
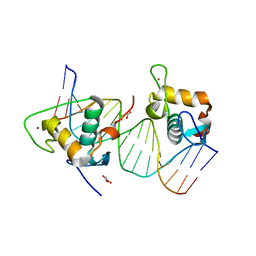 | | Crystal structure of estrogen related receptor-gamma DNA binding domain complexed with Pla2g12b promoter | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*CP*AP*AP*AP*GP*GP*TP*GP*AP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*TP*CP*AP*CP*CP*TP*TP*TP*GP*TP*CP*CP*TP*C)-3'), Estrogen-related receptor gamma, ... | | Authors: | Xu, T, Zhen, X, Liu, J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ERR gamma-DBD undergoes dimerization and conformational rearrangement upon binding to the downstream site of the DR1 element.
Biochem.Biophys.Res.Commun., 656, 2023
|
|
5EH2
 
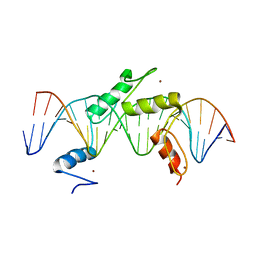 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence III | | Descriptor: | DNA (5'-D(*AP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*CP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*G)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-27 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5EI9
 
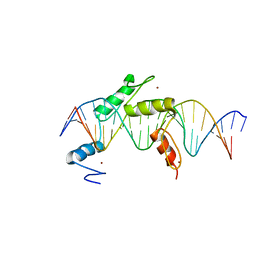 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence I | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*GP*GP*A)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-29 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5EGB
 
 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence II | | Descriptor: | DNA (5'-D(*AP*CP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*GP*G)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5O63
 
 | |
5VBN
 
 | | Crystal Structure of human DNA polymerase epsilon B-subunit in complex with C-terminal domain of catalytic subunit | | Descriptor: | DNA polymerase epsilon catalytic subunit A, DNA polymerase epsilon subunit 2, SULFATE ION, ... | | Authors: | Baranovskiy, A.G, Gu, J, Suwa, Y, Babayeva, N.D, Tahirov, T.H. | | Deposit date: | 2017-03-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the human Pol B-subunit in complex with the C-terminal domain of the catalytic subunit.
J. Biol. Chem., 292, 2017
|
|
3Q4J
 
 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
3Q4K
 
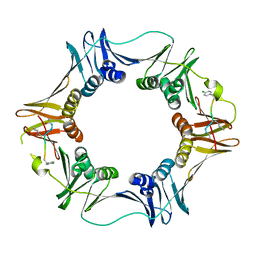 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
6VG2
 
 | |
6VG8
 
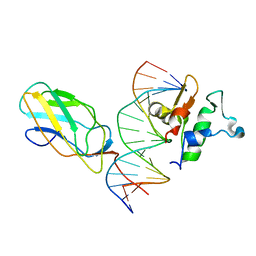 | |
5YI2
 
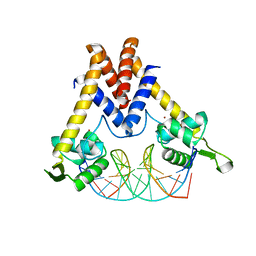 | | Structure of Lactococcus lactis ZitR, wild type in complex with DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*TP*AP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*A)-3'), ZINC ION, Zinc transport transcriptional regulator | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
3WGI
 
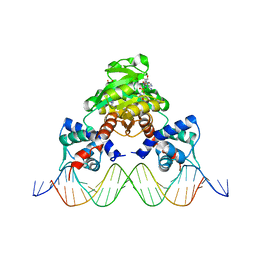 | | Crystal structure of RSP in complex with beta-NAD+ and operator DNA | | Descriptor: | DNA (5'-D(*TP*AP*GP*AP*TP*TP*GP*TP*TP*AP*AP*TP*CP*GP*AP*TP*TP*AP*AP*CP*AP*AP*TP*C)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), Redox-sensing transcriptional repressor rex | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
5X11
 
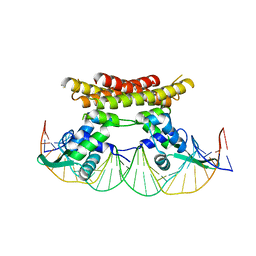 | | Crystal structure of Bacillus subtilis PadR in complex with operator DNA | | Descriptor: | DNA (28-MER), Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
3Q4L
 
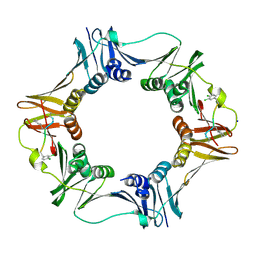 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
5HLF
 
 | |
