7CMU
 
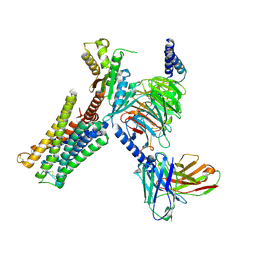 | | Dopamine Receptor D3R-Gi-Pramipexole complex | | Descriptor: | (6S)-N6-propyl-4,5,6,7-tetrahydro-1,3-benzothiazole-2,6-diamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Mao, C, Krumm, B, Zhou, X, Tan, Y, Huang, X.-P, Liu, Y, Shen, D.-D, Jiang, Y, Yu, X, Jiang, H, Melcher, K, Roth, B, Cheng, X, Zhang, Y, Xu, H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human dopamine D3 receptor-G i complexes.
Mol.Cell, 81, 2021
|
|
4WF9
 
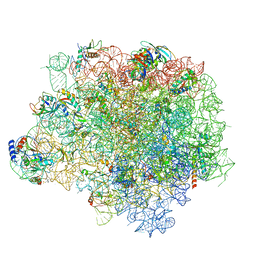 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with telithromycin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S ribosomal RNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A.E. | | Deposit date: | 2014-09-14 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.427 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7CMV
 
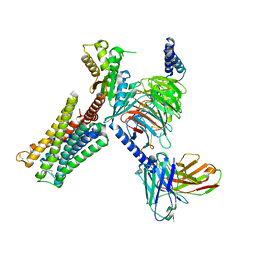 | | Dopamine Receptor D3R-Gi-PD128907 complex | | Descriptor: | (4aR,10bR)-4-propyl-3,4a,5,10b-tetrahydro-2H-chromeno[4,3-b][1,4]oxazin-9-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Mao, C, Krumm, B, Zhou, X, Tan, Y, Huang, X.-P, Liu, Y, Shen, D.-D, Jiang, Y, Yu, X, Jiang, H, Melcher, K, Roth, B, Cheng, X, Zhang, Y, Xu, H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of the human dopamine D3 receptor-G i complexes.
Mol.Cell, 81, 2021
|
|
5LZA
 
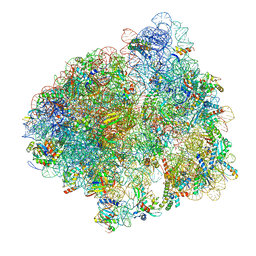 | | Structure of the 70S ribosome with SECIS-mRNA and P-site tRNA (Initial complex, IC) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
7W40
 
 | | Cryo-EM Structure of Human Gastrin Releasing Peptide Receptor in complex with the agonist Bombesin (6-14) [D-Phe6, beta-Ala11, Phe13, Nle14] and Gq heterotrimers | | Descriptor: | Bombesin, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhan, Y, Peng, S, Zhang, H. | | Deposit date: | 2021-11-26 | | Release date: | 2023-02-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of human gastrin-releasing peptide receptors bound to antagonist and agonist for cancer and itch therapy.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8I2G
 
 | | FSHR-Follicle stimulating hormone-compound 716340-Gs complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Duan, J, Xu, P, Yang, J, Ji, Y, Zhang, H, Mao, C, Luan, X, Jiang, Y, Zhang, Y, Zhang, S, Xu, H.E. | | Deposit date: | 2023-01-14 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of hormone and allosteric agonist mediated activation of follicle stimulating hormone receptor.
Nat Commun, 14, 2023
|
|
4RM8
 
 | | Crystal structure of human ezrin in space group P21 | | Descriptor: | Ezrin | | Authors: | Phang, J.M, Harrop, S.J, Davies, R, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2014-10-20 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin.
Biochem. J., 473, 2016
|
|
8IBU
 
 | | Cryo-EM structure of the erythromycin-bound motilin receptor-Gq protein complex | | Descriptor: | ERYTHROMYCIN A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | You, C, Jiang, Y, Xu, H.E, Xu, Y. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural basis for motilin and erythromycin recognition by motilin receptor.
Sci Adv, 9, 2023
|
|
2G9A
 
 | |
8IBV
 
 | | Cryo-EM structure of the motilin-bound motilin receptor-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Jiang, Y, Xu, H.E, You, C, Xu, Y. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for motilin and erythromycin recognition by motilin receptor.
Sci Adv, 9, 2023
|
|
4WFB
 
 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with BC-3205 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S rRNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A.E. | | Deposit date: | 2014-09-14 | | Release date: | 2015-10-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1RIK
 
 | | E6-binding zinc finger (E6apc1) | | Descriptor: | E6apc1 peptide | | Authors: | Liu, Y, Liu, Z, Androphy, E, Chen, J, Baleja, J.D. | | Deposit date: | 2003-11-17 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of helical peptides that inhibit the E6 protein of papillomavirus.
Biochemistry, 43, 2004
|
|
1NPE
 
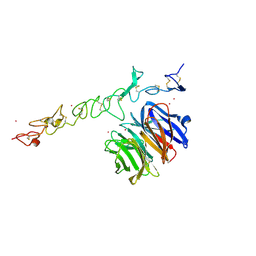 | | Crystal structure of Nidogen/Laminin Complex | | Descriptor: | CADMIUM ION, Laminin gamma-1 chain, nidogen | | Authors: | Takagi, J, Yang, Y.T, Liu, J.-H, Wang, J.-H, Springer, T.A. | | Deposit date: | 2003-01-17 | | Release date: | 2003-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex between nidogen and laminin fragments reveals a paradigmatic
beta-propeller interface
Nature, 424, 2003
|
|
7BTB
 
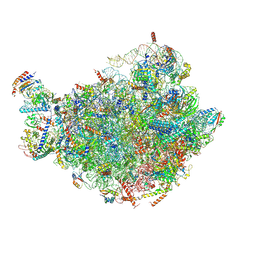 | |
5LZF
 
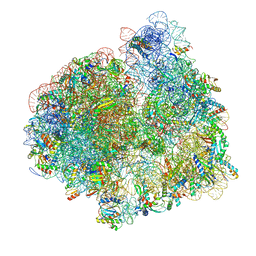 | | Structure of the 70S ribosome with fMetSec-tRNASec in the hybrid pre-translocation state (H) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
1RKS
 
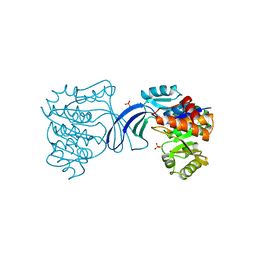 | |
8I02
 
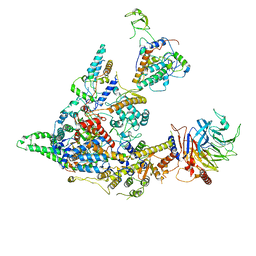 | | Cryo-EM structure of the SIN3S complex from S. pombe | | Descriptor: | Chromatin modification-related protein eaf3, Cph1, Histone deacetylase clr6, ... | | Authors: | Wang, C, Guo, Z, Zhan, X. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Two assembly modes for SIN3 histone deacetylase complexes.
Cell Discov, 9, 2023
|
|
8I03
 
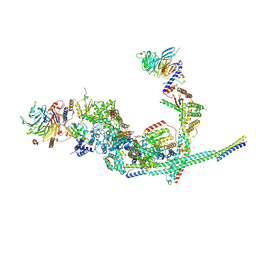 | | Cryo-EM structure of the SIN3L complex from S. pombe | | Descriptor: | Chromatin modification-related protein png2, Histone deacetylase clr6, POTASSIUM ION, ... | | Authors: | Wang, C, Guo, Z, Zhan, X. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Two assembly modes for SIN3 histone deacetylase complexes.
Cell Discov, 9, 2023
|
|
8IA8
 
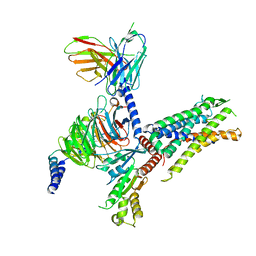 | | Cryo-EM structure of C3aR-Gi-scFv16 bound with E7 peptide | | Descriptor: | ALA-SER-LYS-LEU-GLY-LEU-ALA-ARG, C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liao, Q, Chen, G, Du, Y, Ye, R.D. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Cryo-EM structure of C3aR-Gi-scFv16 bound with E7 peptide
To Be Published
|
|
7C2E
 
 | | GLP-1R-Gs complex structure with a small molecule full agonist | | Descriptor: | 2-[[4-[6-[(4-cyano-2-fluoranyl-phenyl)methoxy]pyridin-2-yl]-3,6-dihydro-2~{H}-pyridin-1-yl]methyl]-3-[[(2~{S})-oxetan-2-yl]methyl]imidazo[4,5-b]pyridine-5-carboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, H, Yuan, D.P, Huang, W, Wenge, Z, Xu, E. | | Deposit date: | 2020-05-07 | | Release date: | 2020-08-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into the activation of GLP-1R by a small molecule agonist.
Cell Res., 30, 2020
|
|
8HJ5
 
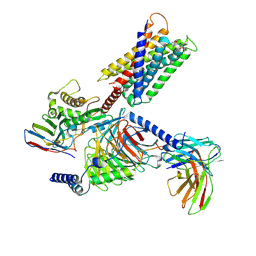 | | Cryo-EM structure of Gq-coupled MRGPRX1 bound with Compound-16 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Gan, B, Yu, L.Y, Ren, R.B. | | Deposit date: | 2022-11-22 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of agonist-induced activation of the human itch receptor MRGPRX1.
Plos Biol., 21, 2023
|
|
8HK5
 
 | | C5aR1-Gi-C5a protein complex | | Descriptor: | C5a anaphylatoxin chemotactic receptor 1, Complement C5, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
4WFN
 
 | | Crystal structure of the large ribosomal subunit (50S) of Deinococcus radiodurans containing a three residue insertion in L22 in complex with erythromycin | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A. | | Deposit date: | 2014-09-16 | | Release date: | 2015-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | The Ribosomal Protein uL22 Modulates the Shape of the Protein Exit Tunnel.
Structure, 25, 2017
|
|
8HK3
 
 | | C3aR-Gi-apo protein complex | | Descriptor: | C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
1O73
 
 | | Tryparedoxin from Trypanosoma brucei | | Descriptor: | TRYPAREDOXIN | | Authors: | Gabrielsen, M, Alphey, M.S, Bond, C.S, Hunter, W.N. | | Deposit date: | 2002-10-23 | | Release date: | 2003-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Tryparedoxins from Crithidia Fasciculata and Trypanosoma Brucei: Photoreduction of the Redox Disulfide Using Synchrotron Radiation and Evidence for a Conformational Switch Implicated in Function
J.Biol.Chem., 278, 2003
|
|
