3EO9
 
 | | Crystal structure the Fab fragment of Efalizumab | | Descriptor: | Efalizumab Fab fragment, heavy chain, light chain | | Authors: | Li, S, Ding, J. | | Deposit date: | 2008-09-26 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efalizumab binding to the LFA-1 alphaL I domain blocks ICAM-1 binding via steric hindrance.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3LZF
 
 | |
6FCZ
 
 | | Model of gC1q-Fc complex based on 7A EM map | | Descriptor: | Complement C1q subcomponent subunit A, Complement C1q subcomponent subunit B, Complement C1q subcomponent subunit C, ... | | Authors: | Ugurlar, D, Howes, S.C, de Kreuk, B.J.K, de Jong, R.N, Beurskens, F.J, Koster, A.J, Parren, P.W.H.I, Sharp, T.H, Gros, P, Koning, R.I. | | Deposit date: | 2017-12-21 | | Release date: | 2018-02-28 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structures of C1-IgG1 provide insights into how danger pattern recognition activates complement.
Science, 359, 2018
|
|
6CE0
 
 | | Crystal structure of a HIV-1 clade B tier-3 isolate H078.14 UFO-BG Env trimer in complex with broadly neutralizing Fabs PGT124 and 35O22 at 4.6 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.602 Å) | | Cite: | HIV-1 vaccine design through minimizing envelope metastability.
Sci Adv, 4, 2018
|
|
6GHG
 
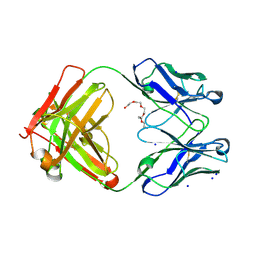 | | Variable heavy - variable light domain and Fab-arm CrossMabs with charged residue exchanges | | Descriptor: | Fab heavy chain, Fab light chain, HEXAETHYLENE GLYCOL, ... | | Authors: | Regula, J, Imhof-Jung, S, Molhoj, M, Benz, J, Ehler, A, Bujotzek, A, Schaefer, W, Klein, C. | | Deposit date: | 2018-05-07 | | Release date: | 2018-09-12 | | Last modified: | 2018-12-05 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Variable heavy-variable light domain and Fab-arm CrossMabs with charged residue exchanges to enforce correct light chain assembly.
Protein Eng. Des. Sel., 31, 2018
|
|
3HKF
 
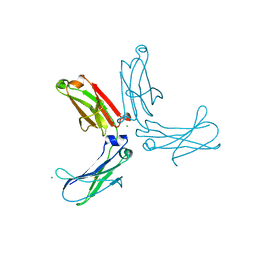 | | Murine unglycosylated IgG Fc fragment | | Descriptor: | CHLORIDE ION, Igh protein, MAGNESIUM ION | | Authors: | Feige, M.J, Nath, S, Catharino, S.R, Weinfurtner, D, Steinbacher, S, Buchner, J. | | Deposit date: | 2009-05-23 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the murine unglycosylated IgG1 Fc fragment
J.Mol.Biol., 391, 2009
|
|
4OUO
 
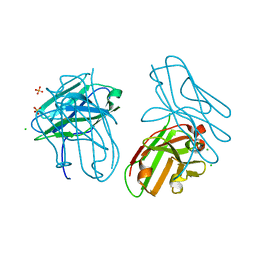 | | anti-Bla g 1 scFv | | Descriptor: | CHLORIDE ION, SULFATE ION, anti Bla g 1 scFv | | Authors: | Mueller, G.A, Ankney, J.A, Glesner, J, Khurana, T, Edwards, L.L, Pedersen, L.C, Perera, L, Slater, J.E, Pomes, A, London, R.E. | | Deposit date: | 2014-02-18 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of an anti-Bla g 1 scFv: Epitope mapping and cross-reactivity.
Mol.Immunol., 59, 2014
|
|
7VAF
 
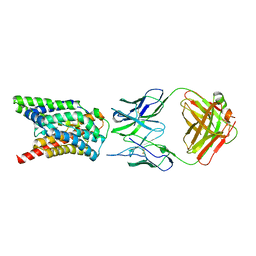 | | Cryo-EM structure of Rat NTCP complexed with YN69202Fab | | Descriptor: | Fab heavy chain from antibody IgG clone number YN69202, Fab light chain from antibody IgG clone number YN69202, Sodium/bile acid cotransporter | | Authors: | Asami, J, Shimizu, T, Ohto, U. | | Deposit date: | 2021-08-29 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structure of the bile acid transporter and HBV receptor NTCP.
Nature, 606, 2022
|
|
7VAG
 
 | |
7VAD
 
 | | Cryo-EM structure of human NTCP complexed with YN69202Fab | | Descriptor: | Fab heavy chain from antibody IgG clone number YN69202, Fab light chain from antibody IgG clone number YN69202, Sodium/bile acid cotransporter | | Authors: | Asami, J, Shimizu, T, Ohto, U. | | Deposit date: | 2021-08-29 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure of the bile acid transporter and HBV receptor NTCP.
Nature, 606, 2022
|
|
7VAE
 
 | | Cryo-EM structure of bovine NTCP complexed with YN69202Fab | | Descriptor: | Fab heavy chain from antibody IgG clone number YN69202, Fab light chain from antibody IgG clone number YN69202, Solute carrier family 10 (Sodium/bile acid cotransporter family), ... | | Authors: | Asami, J, Shimizu, T, Ohto, U. | | Deposit date: | 2021-08-29 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structure of the bile acid transporter and HBV receptor NTCP.
Nature, 606, 2022
|
|
7EH3
 
 | |
1YS5
 
 | | Solution structure of the antigenic domain of GNA1870 of Neisseria meningitidis | | Descriptor: | lipoprotein | | Authors: | Cantini, F, Savino, S, Masignani, V, Pizza, M, Scarselli, M, Swennen, E, Romagnoli, G, Veggi, D, Banci, L, Rappuoli, R. | | Deposit date: | 2005-02-07 | | Release date: | 2006-02-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the immunodominant domain of protective antigen GNA1870 of Neisseria meningitidis
J.Biol.Chem., 281, 2006
|
|
7F5H
 
 | | The crystal structure of RBD-Nanobody complex, DL28 (SC4) | | Descriptor: | GLYCEROL, Nanobody DL28, PHOSPHATE ION, ... | | Authors: | Luo, Z.P, Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of a Neutralizing Nanobody With Broad Activity Against SARS-CoV-2 Variants.
Front Microbiol, 13, 2022
|
|
8DE6
 
 | | Oligomeric C9 in complex with aE11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement component C9, ... | | Authors: | Bayly-Jones, C. | | Deposit date: | 2022-06-20 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The neoepitope of the complement C5b-9 Membrane Attack Complex is formed by proximity of adjacent ancillary regions of C9.
Commun Biol, 6, 2023
|
|
8D01
 
 | |
7WSI
 
 | |
7WLW
 
 | | X-ray structure of thermostabilized Drosophila dopamine transporter with GABA transporter1-like substitutions in the binding site, in complex with SKF89976a | | Descriptor: | (3S)-1-(4,4-diphenylbut-3-enyl)piperidine-3-carboxylic acid, Antibody Fragment heavy chain, Antibody fragment light chain, ... | | Authors: | Penmatsa, A, Joseph, D. | | Deposit date: | 2022-01-13 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into GABA transport inhibition using an engineered neurotransmitter transporter.
Embo J., 41, 2022
|
|
8DS7
 
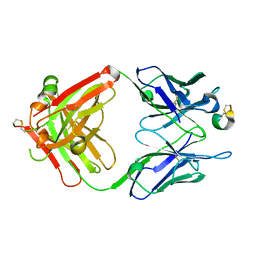 | |
5LHP
 
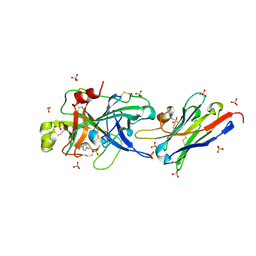 | | The p-aminobenzamidine active site inhibited catalytic domain of murine urokinase-type plasminogen activator in complex with the allosteric inhibitory nanobody Nb7 | | Descriptor: | 1,2-ETHANEDIOL, Camelid-Derived Antibody Fragment, P-AMINO BENZAMIDINE, ... | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P.J, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
6UGY
 
 | | Crystal structure of the Fc fragment of anti-TNFa antibody infliximab (Remicade) in a primative orthorhombic crystal form, Lot C | | Descriptor: | ACETATE ION, Remicade Fc, ZINC ION, ... | | Authors: | Mayclin, S.J, Edwards, T.E. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
6U9S
 
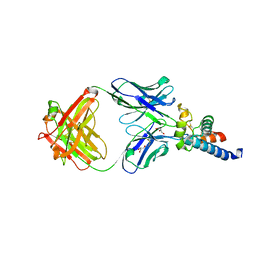 | | Crystal structure of human CD81 large extracellular loop in complex with 5A6 Fab | | Descriptor: | 5A6 FAB Heavy Chain, 5A6 FAB Light Chain, CD81 antigen, ... | | Authors: | Susa, K.J, Seegar, T.C.M, Blacklow, S.C.B, Kruse, A.C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A dynamic interaction between CD19 and the tetraspanin CD81 controls B cell co-receptor trafficking.
Elife, 9, 2020
|
|
5LGK
 
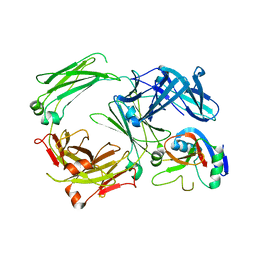 | | Crystal structure of the human IgE-Fc bound to its B cell receptor derCD23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ig epsilon chain C region, Low affinity immunoglobulin epsilon Fc receptor, ... | | Authors: | Dhaliwal, B, Pang, M.O.Y, Sutton, B.J. | | Deposit date: | 2016-07-07 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | IgE binds asymmetrically to its B cell receptor CD23.
Sci Rep, 7, 2017
|
|
5MOJ
 
 | | Crystal structure of IgE-Fc epsilon 3-4 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ig epsilon chain C region, ... | | Authors: | Dore, K.A, Davies, A.M, Drinkwater, N, Beavil, A.J, MacDonnell, J.M, Sutton, B.J. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Thermal sensitivity and flexibility of the C epsilon 3 domains in immunoglobulin E.
Biochim. Biophys. Acta, 1865, 2017
|
|
5MOI
 
 | | Crystal structure of human IgE-Fc epsilon 3-4 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ig epsilon chain C region, ... | | Authors: | Dore, K.A, Davies, A.M, Drinkwater, N, Beavil, A.J, McDonnell, J.M, Sutton, B.J. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thermal sensitivity and flexibility of the C epsilon 3 domains in immunoglobulin E.
Biochim. Biophys. Acta, 1865, 2017
|
|
