5FYD
 
 | | Structural and biochemical insights into 7beta-hydroxysteroid dehydrogenase stereoselectivity | | Descriptor: | GLYCEROL, OXIDOREDUCTASE, SHORT CHAIN DEHYDROGENASE/REDUCTASE FAMILY PROTEIN | | Authors: | Savino, S, Ferrandi, E, Forneris, F, Rovida, S, Riva, S, Monti, D, Mattevi, A. | | Deposit date: | 2016-03-07 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Insights Into 7Beta-Hydroxysteroid Dehydrogenase Stereoselectivity.
Proteins, 84, 2016
|
|
6QU0
 
 | | Structure of azoreductase from Bacillus sp. A01 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase, GLYCEROL, ... | | Authors: | Savino, S, Fraaije, M.W. | | Deposit date: | 2019-02-26 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic and Crystallographic Studies of Azoreductase AzoA fromBacillus wakoensisA01.
Acs Chem.Biol., 15, 2020
|
|
5MQ6
 
 | | Polycyclic Ketone Monooxygenase from the Thermophilic Fungus Thermothelomyces thermophila | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Savino, S, Furst, M.J.L.J, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2016-12-20 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Polycyclic Ketone Monooxygenase from the Thermophilic Fungus Thermothelomyces thermophila: A Structurally Distinct Biocatalyst for Bulky Substrates.
J. Am. Chem. Soc., 139, 2017
|
|
6Y0R
 
 | | Chitooligosaccharide oxidase | | Descriptor: | Chitooligosaccharide oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Savino, S, Fraaije, M.W. | | Deposit date: | 2020-02-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Analysis of the structure and substrate scope of chitooligosaccharide oxidase reveals high affinity for C2-modified glucosamines.
Febs Lett., 594, 2020
|
|
6H0P
 
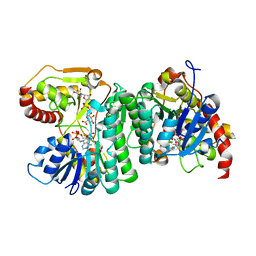 | |
6H0N
 
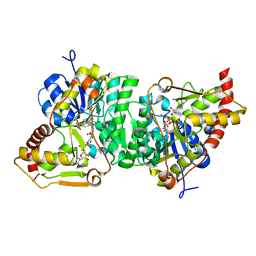 | |
8ATE
 
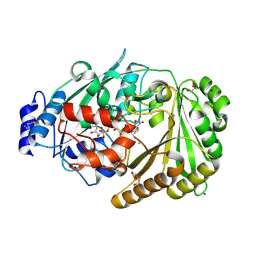 | | Galacturonic acid oxidase from Citrus sinensis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boverio, A, Savino, S, Fraaije, M.W, Loncar, N. | | Deposit date: | 2022-08-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and Structural Characterization of a Uronic Acid Oxidase from Citrus sinensis
Chemcatchem, 2023
|
|
7OG2
 
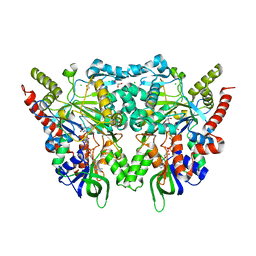 | |
5AJU
 
 | |
5AJT
 
 | | Crystal structure of ligand-free phosphoribohydrolase lonely guy from Claviceps purpurea | | Descriptor: | 1,2-ETHANEDIOL, D(-)-TARTARIC ACID, PHOSPHORIBOHYDROLASE LONELY GUY | | Authors: | Dzurova, L, Savino, S, Forneris, F. | | Deposit date: | 2015-02-27 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The three-dimensional structure of "Lonely Guy" from Claviceps purpurea provides insights into the phosphoribohydrolase function of Rossmann fold-containing lysine decarboxylase-like proteins.
Proteins, 83, 2015
|
|
1YS5
 
 | | Solution structure of the antigenic domain of GNA1870 of Neisseria meningitidis | | Descriptor: | lipoprotein | | Authors: | Cantini, F, Savino, S, Masignani, V, Pizza, M, Scarselli, M, Swennen, E, Romagnoli, G, Veggi, D, Banci, L, Rappuoli, R. | | Deposit date: | 2005-02-07 | | Release date: | 2006-02-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the immunodominant domain of protective antigen GNA1870 of Neisseria meningitidis
J.Biol.Chem., 281, 2006
|
|
7ZCB
 
 | | Human Pikachurin/EGFLAM N-terminal Fibronectin-III (1-2) domains | | Descriptor: | CHLORIDE ION, Pikachurin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Pantalone, S, Savino, S, Viti, L.V, Forneris, F. | | Deposit date: | 2022-03-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179.
Sci.Signal., 16, 2023
|
|
2KC0
 
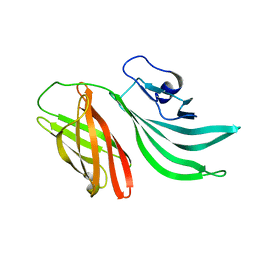 | | Solution structure of the factor H binding protein | | Descriptor: | lipoprotein | | Authors: | Cantini, F, Veggi, D, Dragonetti, S, Savino, S, Scarselli, M, Romagnoli, G, Pizza, M, Banci, L, Rappuoli, R. | | Deposit date: | 2008-12-13 | | Release date: | 2009-02-17 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Factor H-binding Protein, a Survival Factor and Protective Antigen of Neisseria meningitidis
J.Biol.Chem., 284, 2009
|
|
6ZLD
 
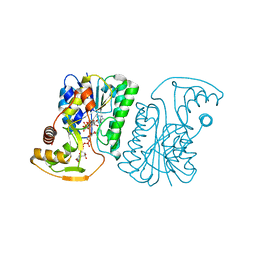 | | Crystal Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Glucuronic acid and NAD | | Descriptor: | Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Iacovino, L.G, Savino, S, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
6ZLL
 
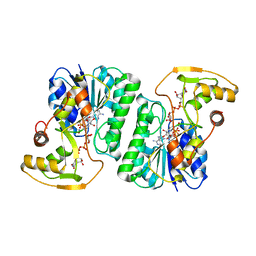 | | Crystal Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Galacturonic acid and NAD | | Descriptor: | (2S,3R,4S,5R,6R)-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4,5-trihydroxy-oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Iacovino, L.G, Savino, S, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
