3T32
 
 | | Crystal structure of a putative C-S lyase from Bacillus anthracis | | Descriptor: | Aminotransferase, class I/II | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Peterson, S.N, Porebski, P, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-24 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative C-S lyase from Bacillus anthracis
TO BE PUBLISHED
|
|
9J7H
 
 | | Crystal structure of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from Providencia alcalifaciens complexed with quinic acid | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Phospho-2-dehydro-3-deoxyheptonate aldolase | | Authors: | Jangid, K, Mahto, J.K, Kumar, K.A, Kumar, P. | | Deposit date: | 2024-08-19 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural and biochemical analyses reveal quinic acid inhibits DAHP synthase a key player in shikimate pathway.
Arch.Biochem.Biophys., 763, 2025
|
|
9J7S
 
 | | Crystal structure of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from Providencia alcalifaciens complexed with Phe | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHENYLALANINE, Phospho-2-dehydro-3-deoxyheptonate aldolase | | Authors: | Jangid, K, Mahto, J.K, Kumar, K.A, Kumar, P. | | Deposit date: | 2024-08-19 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses reveal quinic acid inhibits DAHP synthase a key player in shikimate pathway.
Arch.Biochem.Biophys., 763, 2025
|
|
9J19
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor minocycline | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, 3C-like proteinase nsp5 | | Authors: | Singh, A, Jangid, K, Dhaka, P, Tomar, S, Kumar, P. | | Deposit date: | 2024-08-04 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mechanistic Insights into the Main Protease (Mpro) Dimer Interface Destabilization Inhibitor: Unveiling New Therapeutic Avenues against SARS-CoV-2.
Biochemistry, 64, 2025
|
|
9IN1
 
 | |
3NXF
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | Descriptor: | Retro-Aldolase, SULFATE ION | | Authors: | Althoff, E.A, Jiang, L, Wang, L, Lassila, J.K, Moody, J, Bolduc, J, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | Deposit date: | 2010-07-13 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
3O2E
 
 | |
5DOW
 
 | |
3NWV
 
 | | Human cytochrome c G41S | | Descriptor: | Cytochrome c, HEME C | | Authors: | Fagerlund, R.D, Wilbanks, S.M. | | Deposit date: | 2010-07-11 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Proapoptotic G41S Mutation to Human Cytochrome c Alters the Heme Electronic Structure and Increases the Electron Self-Exchange Rate.
J.Am.Chem.Soc., 133, 2011
|
|
7SAP
 
 | | The CTI-homolog pacifastin | | Descriptor: | GLYCEROL, Serine protease inhibitor I/II-like Protein | | Authors: | Gewe, M.M, Strong, R.K. | | Deposit date: | 2021-09-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Ex silico engineering of cystine-dense peptides yielding a potent bispecific T cell engager.
Sci Transl Med, 14, 2022
|
|
6EVV
 
 | | X-ray structure of the complex between human alpha thrombin and NU172, a duplex/quadruplex 26-mer DNA aptamer, in the presence of potassium ions. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Troisi, R, Russo Krauss, I, Sica, F. | | Deposit date: | 2017-11-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer.
Nucleic Acids Res., 46, 2018
|
|
9BOH
 
 | | Room-temperature X-ray structure of Thermus Thermophilus serine hydroxymethyltransferase (SHMT) with PLP-glycine external aldimine and 5-formyltetrahydrofolate (folinic acid) | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)glycine, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SULFATE ION, ... | | Authors: | Drago, V.N, Kovalevsky, A. | | Deposit date: | 2024-05-03 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Universality of critical active site glutamate as an acid-base catalyst in serine hydroxymethyltransferase function.
Chem Sci, 15, 2024
|
|
9BOW
 
 | | X-ray structure of Thermus thermophilus serine hydroxymethyltransferase with PLP-L-Ser external aldimine and 5-formyltetrahydrofolate (folinic acid) | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SERINE, ... | | Authors: | Drago, V.N, Kovalevsky, A. | | Deposit date: | 2024-05-06 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Universality of critical active site glutamate as an acid-base catalyst in serine hydroxymethyltransferase function.
Chem Sci, 15, 2024
|
|
9BPE
 
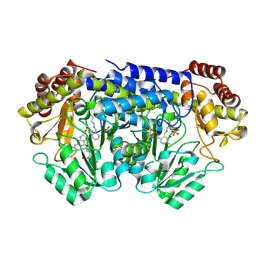 | | Joint X-ray/neutron structure of Thermus thermophilus serine hydroxymethyltransferase (TthSHMT) in internal aldimine state and folinic acid bound | | Descriptor: | ACETATE ION, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SULFATE ION, ... | | Authors: | Drago, V.N, Kovalevsky, A. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-28 | | Method: | NEUTRON DIFFRACTION (2.3 Å), X-RAY DIFFRACTION | | Cite: | Universality of critical active site glutamate as an acid-base catalyst in serine hydroxymethyltransferase function.
Chem Sci, 15, 2024
|
|
7T0F
 
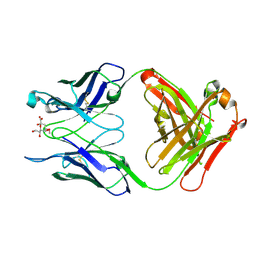 | |
7ST8
 
 | | Crystal structure of 7H2.2 Fab in complex with SAS1B C-terminal region | | Descriptor: | 7H2.2 Fab Heavy Chain, 7H2.2 Fab Light Chain, Astacin-like metalloendopeptidase | | Authors: | Legg, M.S.G, Evans, S.V. | | Deposit date: | 2021-11-12 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Monoclonal antibody 7H2.2 binds the C-terminus of the cancer-oocyte antigen SAS1B through the hydrophilic face of a conserved amphipathic helix corresponding to one of only two regions predicted to be ordered
Acta Crystallogr.,Sect.D, 78, 2022
|
|
7SOH
 
 | |
3Q8T
 
 | |
7SGQ
 
 | |
7SLT
 
 | |
7SND
 
 | | Pacifastin related protease inhibitors | | Descriptor: | GLYCEROL, PHOSPHATE ION, Pacifastin-related peptide | | Authors: | Gewe, M.M, Strong, R.K. | | Deposit date: | 2021-10-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Ex silico engineering of cystine-dense peptides yielding a potent bispecific T cell engager.
Sci Transl Med, 14, 2022
|
|
7SNC
 
 | | Pacifastin related protease inhibitors | | Descriptor: | Protease inhibitor | | Authors: | Gewe, M.M, Strong, R.K. | | Deposit date: | 2021-10-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ex silico engineering of cystine-dense peptides yielding a potent bispecific T cell engager.
Sci Transl Med, 14, 2022
|
|
3PMB
 
 | |
7TOB
 
 | | Crystal structure of the SARS-CoV-2 Omicron main protease (Mpro) in complex with inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Sacco, M.D, Wang, J, Chen, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition.
Cell Res., 32, 2022
|
|
3PM6
 
 | |
