1QME
 
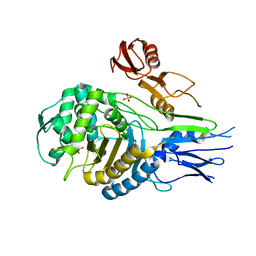 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2X, SULFATE ION | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Penicillin-Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance.
J.Mol.Biol., 299, 2000
|
|
1QMF
 
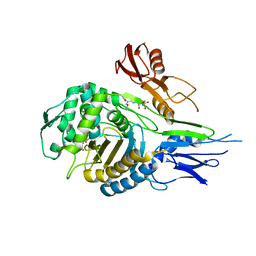 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) ACYL-ENZYME COMPLEX | | Descriptor: | 2-[CARBOXY-(2-FURAN-2-YL-2-METHOXYIMINO-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, CEFUROXIME (OCT-3-ENE FORM), PENICILLIN-BINDING PROTEIN 2X | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the Penicillin Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance
J.Mol.Biol., 299, 2000
|
|
1I4U
 
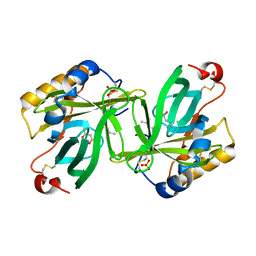 | | THE C1 SUBUNIT OF ALPHA-CRUSTACYANIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CRUSTACYANIN, SULFATE ION | | Authors: | Gordon, E.J, Leonard, G.A, McSweeney, S, Zagalsky, P.F. | | Deposit date: | 2001-02-23 | | Release date: | 2001-09-19 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The C1 subunit of alpha-crustacyanin: the de novo phasing of the crystal structure of a 40 kDa homodimeric protein using the anomalous scattering from S atoms combined with direct methods.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1E8C
 
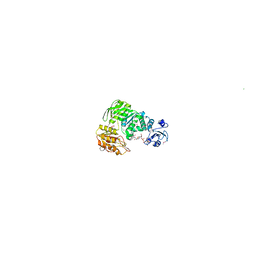 | | Structure of MurE the UDP-N-acetylmuramyl tripeptide synthetase from E. coli | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CHLORIDE ION, UDP-N-ACETYLMURAMOYLALANYL-D-GLUTAMATE--2,6-DIAMINOPIMELATE LIGASE, ... | | Authors: | Gordon, E.J, Chantala, L, Dideberg, O. | | Deposit date: | 2000-09-19 | | Release date: | 2001-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Udp-N-Acetylmuramoyl-L-Alanyl-D-Glutamate: Meso-Diaminopimelate Ligase from Escherichia Coli
J.Biol.Chem., 276, 2001
|
|
3KBY
 
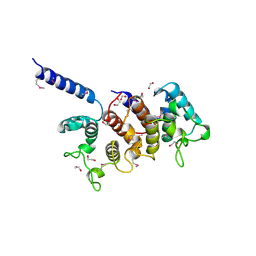 | | Crystal structure of hypothetical protein from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein | | Authors: | Lam, R, Thompson, C.M, Battaile, K.P, Romanov, V, Kisselman, G, Gordon, E, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-10-20 | | Release date: | 2010-10-20 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hypothetical protein from Staphylococcus aureus
To be Published
|
|
3L44
 
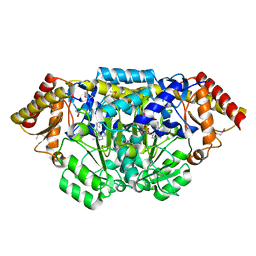 | | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase | | Descriptor: | Glutamate-1-semialdehyde 2,1-aminomutase 1 | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase
TO BE PUBLISHED
|
|
1GQ1
 
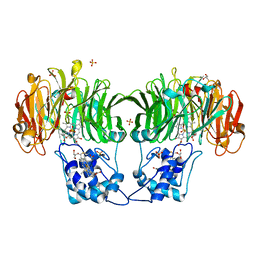 | | CYTOCHROME CD1 NITRITE REDUCTASE, Y25S mutant, OXIDISED FORM | | Descriptor: | CYTOCHROME CD1 NITRITE REDUCTASE, GLYCEROL, HEME C, ... | | Authors: | Sjogren, T, Gordon, E.H.J, Lofqvist, M, Richter, C.D, Hajdu, J, Ferguson, S.J. | | Deposit date: | 2001-11-19 | | Release date: | 2002-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and Kinetic Properties of Paracoccus Pantotrophus Cytochrome Cd1 Nitrite Reductase with the D1 Heme Active Site Ligand Tyrosine 25 Replaced by Serine
J.Biol.Chem., 278, 2003
|
|
4EHJ
 
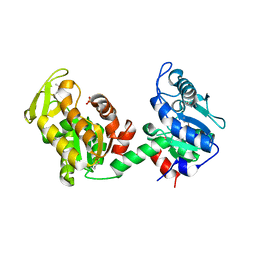 | | An X-ray Structure of a Putative Phosphogylcerate Kinase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Phosphoglycerate kinase, SULFATE ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Gordon, E, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-05-09 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | An X-ray Structure of a Putative Phosphogylcerate Kinase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
7O19
 
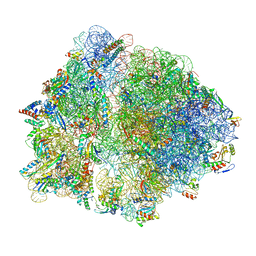 | | Cryo-EM structure of an Escherichia coli TnaC-ribosome complex stalled in response to L-tryptophan | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | van der Stel, A.X, Gordon, E.R, Sengupta, A, Martinez, A.K, Klepacki, D, Perry, T.N, Herrero del Valle, A, Vazquez-Laslop, N, Sachs, M.S, Cruz-Vera, L.R, Innis, C.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-09-01 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling.
Nat Commun, 12, 2021
|
|
7O1A
 
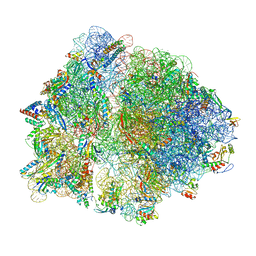 | | Cryo-EM structure of an Escherichia coli TnaC(R23F)-ribosome complex stalled in response to L-tryptophan | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | van der Stel, A.X, Gordon, E.R, Sengupta, A, Martinez, A.K, Klepacki, D, Perry, T.N, Herrero del Valle, A, Vazquez-Laslop, N, Sachs, M.S, Cruz-Vera, L.R, Innis, C.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-09-01 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling.
Nat Commun, 12, 2021
|
|
7O1C
 
 | | Cryo-EM structure of an Escherichia coli TnaC(R23F)-ribosome-RF2 complex stalled in response to L-tryptophan | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | van der Stel, A.X, Gordon, E.R, Sengupta, A, Martinez, A.K, Klepacki, D, Perry, T.N, Herrero del Valle, A, Vazquez-Laslop, N, Sachs, M.S, Cruz-Vera, L.R, Innis, C.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-09-01 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling.
Nat Commun, 12, 2021
|
|
3KWL
 
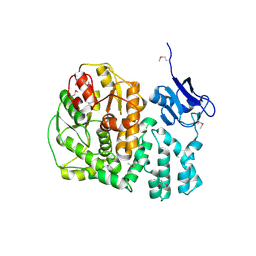 | | Crystal structure of a hypothetical protein from Helicobacter pylori | | Descriptor: | uncharacterized protein | | Authors: | Lam, R, Thompson, C.M, Vodsedalek, J, Lam, K, Romanov, V, Battaile, K.P, Beletskaya, I, Gordon, E, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-12-01 | | Release date: | 2010-12-01 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of a hypothetical protein from Helicobacter pylori
To be Published
|
|
2CE4
 
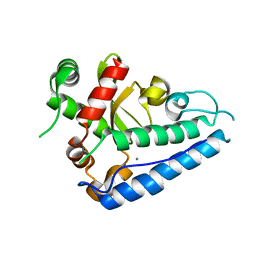 | | Manganese Superoxide Dismutase (Mn-SOD) from Deinococcus radiodurans | | Descriptor: | MANGANESE (II) ION, SUPEROXIDE DISMUTASE [MN] | | Authors: | Dennis, R, Micossi, E, McCarthy, J, Moe, E, Gordon, E, Leonard, G, McSweeney, S. | | Deposit date: | 2006-02-03 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the manganese superoxide dismutase from Deinococcus radiodurans in two crystal forms.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 62, 2006
|
|
2CDY
 
 | | Manganese Superoxide Dismutase (Mn-SOD) from Deinococcus radiodurans | | Descriptor: | MANGANESE (II) ION, SUPEROXIDE DISMUTASE [MN] | | Authors: | Dennis, R, Micossi, E, McCarthy, J, Moe, E, Gordon, E, Leonard, G, McSweeney, S. | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the manganese superoxide dismutase from Deinococcus radiodurans in two crystal forms.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 62, 2006
|
|
3LYL
 
 | | Structure of 3-oxoacyl-acylcarrier protein reductase, FabG from Francisella tularensis | | Descriptor: | 3-oxoacyl-(Acyl-carrier-protein) reductase | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-27 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of 3-oxoacyl-acylcarrier protein reductase, FabG from Francisella tularensis
TO BE PUBLISHED
|
|
3Q7H
 
 | | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, DI(HYDROXYETHYL)ETHER | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii
To be Published
|
|
2FDP
 
 | | Crystal structure of beta-secretase complexed with an amino-ethylene inhibitor | | Descriptor: | Beta-secretase 1, N1-((2S,3S,5R)-3-AMINO-6-(4-FLUOROPHENYLAMINO)-5-METHYL-6-OXO-1-PHENYLHEXAN-2-YL)-N3,N3-DIPROPYLISOPHTHALAMIDE | | Authors: | Yang, W, Lu, W, Lu, Y, Zhong, M, Sun, J, Thomas, A.E, Wilkinson, J.M, Fucini, R.V, Lam, M, Randal, M, Shi, X.P, Jacobs, J.W, McDowell, R.S, Gordon, E.M, Ballinger, M.D. | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminoethylenes: a tetrahedral intermediate isostere yielding potent inhibitors of the aspartyl protease BACE-1.
J.Med.Chem., 49, 2006
|
|
3KUX
 
 | | Structure of the YPO2259 putative oxidoreductase from Yersinia pestis | | Descriptor: | CHLORIDE ION, Putative oxidoreductase | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Kwon, K, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the YPO2259 putative oxidoreductase from Yersinia pestis
To be Published
|
|
3KOM
 
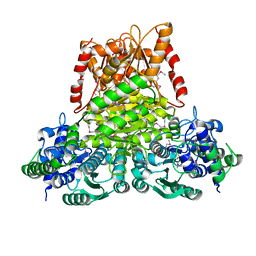 | | Crystal structure of apo transketolase from Francisella tularensis | | Descriptor: | Transketolase | | Authors: | Anderson, S.M, Wawrzak, Z, Skarina, T, Gordon, E, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-11-13 | | Release date: | 2009-12-01 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of apo transketolase from Francisella tularensis
TO BE PUBLISHED
|
|
2VF7
 
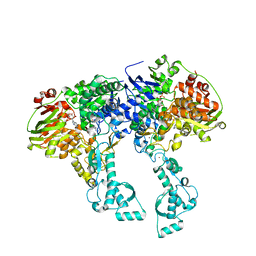 | | Crystal structure of UvrA2 from Deinococcus radiodurans | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EXCINUCLEASE ABC, SUBUNIT A., ... | | Authors: | Timmins, J, Gordon, E, Caria, S, Leonard, G, Kuo, M.S, Monchois, V, McSweeney, S. | | Deposit date: | 2007-10-31 | | Release date: | 2008-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational analyses of Deinococcus radiodurans UvrA2 provide insight into DNA binding and damage recognition by UvrAs.
Structure, 17, 2009
|
|
2VF8
 
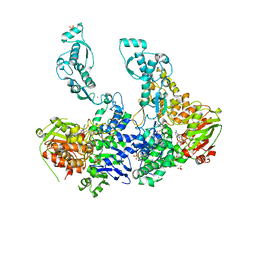 | | Crystal structure of UvrA2 from Deinococcus radiodurans | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EXCINUCLEASE ABC SUBUNIT A, PHOSPHATE ION, ... | | Authors: | Timmins, J, Gordon, E, Caria, S, Leonard, G, Kuo, M.S, Monchois, V, McSweeney, S. | | Deposit date: | 2007-10-31 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Mutational Analyses of Deinococcus Radiodurans Uvra2 Provide Insight Into DNA Binding and Damage Recognition by Uvras.
Structure, 17, 2009
|
|
3T32
 
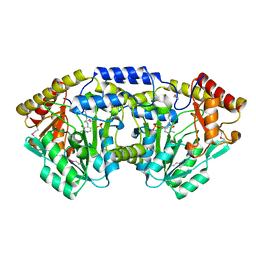 | | Crystal structure of a putative C-S lyase from Bacillus anthracis | | Descriptor: | Aminotransferase, class I/II | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Peterson, S.N, Porebski, P, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-24 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative C-S lyase from Bacillus anthracis
TO BE PUBLISHED
|
|
3OF5
 
 | | Crystal Structure of a Dethiobiotin Synthetase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | ACETATE ION, Dethiobiotin synthetase, SODIUM ION | | Authors: | Brunzelle, J.S, Skarina, T, Gordon, E, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-13 | | Release date: | 2011-02-02 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of a Dethiobiotin Synthetase from Francisella tularensis subsp. tularensis SCHU S4
TO BE PUBLISHED
|
|
3P7M
 
 | | Structure of putative lactate dehydrogenase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Malate dehydrogenase, PHOSPHATE ION | | Authors: | Osinski, T, Cymborowski, M, Zimmerman, M.D, Gordon, E, Grimshaw, S, Skarina, T, Chruszcz, M, Savchenko, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-12 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of putative lactate dehydrogenase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
3IJR
 
 | | 2.05 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' in complex with NAD+ | | Descriptor: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase, ... | | Authors: | Halavaty, A.S, Minasov, G, Skarina, T, Onopriyenko, O, Gordon, E, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' in complex with NAD+
To be Published
|
|
