6MHA
 
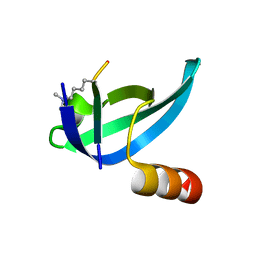 | | dHP1 Chromodomain Y24W variant bound to histone H3 peptide containing trimethyllysine | | Descriptor: | Heterochromatin protein 1, histone H3 peptide containing trimethyllysine, H3K9me3 | | Authors: | Brustad, E.M, Krone, M.W, Albanese, K.I, Waters, M.L. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Thermodynamic consequences of Tyr to Trp mutations in the cation-pi-mediated binding of trimethyllysine by the HP1 chromodomain
Chem Sci, 11, 2020
|
|
8RQU
 
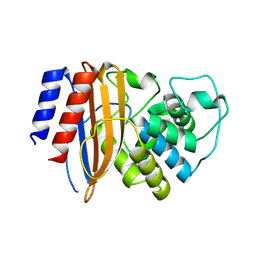 | | Structure of TEM1 beta-lactamase variant 70.a | | Descriptor: | Beta-lactamase TEM-1, MAGNESIUM ION | | Authors: | Napier, E, Fram, B.F, Gauthier, N.P, Sander, C, Khan, A.R. | | Deposit date: | 2024-01-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Simultaneous enhancement of multiple functional properties using evolution-informed protein design.
Nat Commun, 15, 2024
|
|
9LCR
 
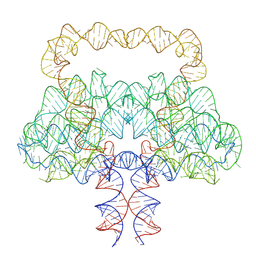 | | Clostridium botulinum OLE RNA dimer | | Descriptor: | MAGNESIUM ION, RNA (578-MER) | | Authors: | Jia, X.Y, Wang, L, Su, Z.M. | | Deposit date: | 2025-01-05 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM reveals mechanisms of natural RNA multivalency.
Science, 388, 2025
|
|
9N2B
 
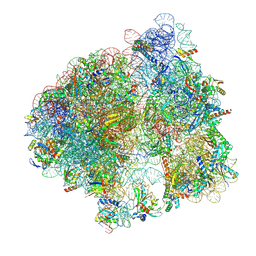 | | Impacts of ribosomal RNA sequence variation on gene expression and phenotype: Cryo-EM structure of the rrsB ribosome (BBB-70S) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA (rRNA) from the rrnB operon, 23S ribosomal RNA (rRNA) from the rrnB operon, ... | | Authors: | Welfer, G.A, Brady, R.A, Natchiar, S.K, Watson, Z.L, Rundlet, E.J, Alejo, J.L, Singh, A.P, Mishra, N.K, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2025-01-28 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Impacts of ribosomal RNA sequence variation on gene expression and phenotype.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 380, 2025
|
|
9N2C
 
 | | Impacts of ribosomal RNA sequence variation on gene expression and phenotype: Cryo-EM structure of the rrsH ribosome (HBB-70S) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA (rRNA) from the rrnH operon, 23S ribosomal RNA (rRNA) from the rrnB operon, ... | | Authors: | Welfer, G.A, Brady, R.A, Natchiar, S.K, Watson, Z.L, Rundlet, E.J, Alejo, J.L, Singh, A.P, Mishra, N.K, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2025-01-28 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Impacts of ribosomal RNA sequence variation on gene expression and phenotype.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 380, 2025
|
|
5IJC
 
 | | The crystal structure of mouse TLR4/MD-2/neoseptin-3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7R81
 
 | | Structure of the translating Neurospora crassa ribosome arrested by cycloheximide | | Descriptor: | 18S rRNA, 26S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Shen, L, Su, Z, Yang, K, Wu, C, Becker, T, Bell-Pedersen, D, Zhang, J, Sachs, M.S. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the translating Neurospora ribosome arrested by cycloheximide
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4JLL
 
 | | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, OSH55.4_H1 covalently bound with FP-alkyne, Northeast Structural Genomics Consortium (NESG) Target OR273 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Tong, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ETK
 
 | | Crystal Structure of E6A/L130D/A155H variant of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR186 | | Descriptor: | De novo designed serine hydrolase, SODIUM ION | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ETJ
 
 | | Crystal Structure of E6H variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR185 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
9J3T
 
 | |
5VHG
 
 | | Crystal structure of pentad mutant GAPR-1 | | Descriptor: | Golgi-associated plant pathogenesis-related protein 1, SULFATE ION | | Authors: | Li, Y, Zhao, Y, Su, M, Chakravarthy, S, Colbert, C.L, Levine, B, Sinha, S.C. | | Deposit date: | 2017-04-13 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural insights into the interaction of the conserved mammalian proteins GAPR-1 and Beclin 1, a key autophagy protein.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
9J3R
 
 | |
6J76
 
 | | Structure of 3,6-anhydro-L-galactose Dehydrogenase in Complex with NAP | | Descriptor: | Aldehyde dehydrogenase A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, P.Y, Wang, Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-01-17 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | 3,6-Anhydro-L-Galactose Dehydrogenase VvAHGD is a Member of a New Aldehyde Dehydrogenase Family and Catalyzes by a Novel Mechanism with Conformational Switch of Two Catalytic Residues Cysteine 282 and Glutamate 248.
J.Mol.Biol., 432, 2020
|
|
6J75
 
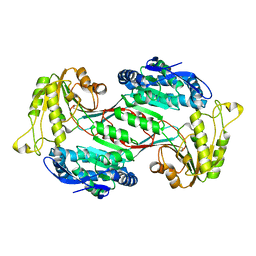 | | Structure of 3,6-anhydro-L-galactose Dehydrogenase | | Descriptor: | Aldehyde dehydrogenase A | | Authors: | Li, P.Y, Wang, Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-01-17 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | 3,6-Anhydro-L-Galactose Dehydrogenase VvAHGD is a Member of a New Aldehyde Dehydrogenase Family and Catalyzes by a Novel Mechanism with Conformational Switch of Two Catalytic Residues Cysteine 282 and Glutamate 248.
J.Mol.Biol., 432, 2020
|
|
7WBQ
 
 | |
9KPH
 
 | |
9KPO
 
 | |
5C2O
 
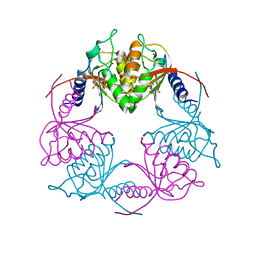 | | Crystal structure of Streptococcus mutans Deoxycytidylate Deaminase complexed with dTTP | | Descriptor: | MAGNESIUM ION, Putative deoxycytidylate deaminase, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Li, Y.H, Gao, Z.Q, Hou, H.F, Dong, Y.H. | | Deposit date: | 2015-06-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of the allosteric regulation of Streptococcus mutans 2'-deoxycytidylate deaminase.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
9ISV
 
 | |
7WBP
 
 | |
5YPS
 
 | | The structural basis of histone chaperoneVps75 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | Deposit date: | 2017-11-03 | | Release date: | 2018-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
5ZB5
 
 | | The structural basis of histone chaperoneVps75 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NAP family histone chaperone vps75 | | Authors: | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | Deposit date: | 2018-02-09 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
5SY5
 
 | | Crystal Structure of the Heterodimeric NPAS1-ARNT Complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Neuronal PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Potluri, N, Kim, Y, Rastinejad, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors.
Elife, 5, 2016
|
|
5SY7
 
 | | Crystal Structure of the Heterodimeric NPAS3-ARNT Complex with HRE DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*CP*AP*CP*GP*AP*CP*CP*CP*GP*CP*AP*CP*GP*TP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*GP*CP*GP*TP*AP*CP*GP*TP*GP*CP*GP*GP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Wu, D, Su, X, Potluri, N, Kim, Y, Rastinejad, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors.
Elife, 5, 2016
|
|
